Kraken 2 tutorial
Here I will try to see what kind of bacteria and viruses lie within the RNAseq of a clade of nematodes. The clade is the Tylenchida, a clade with diverse lifestyles, but most interestingly, lots of parasites.
Kraken has a lot of standardized databases that can be downloaded, though the more species/clades you include, the longer it takes to make the kraken database.
Lots of great information can be had at the Kraken2 wiki
Step 1: Build an appropriate kraken2 database
Note that this is a slight hack to the normal database build, but allowed the build
Modify download_taxonomy.sh
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
#download taxonomy data from ncbi
module load GIF/kraken2
#kraken2-build --download-taxonomy --db NematodeViral
#This should have worked, but NCBI removed a couple files from their ftp site (est and gss)
Had to modify the download_taxonomy.sh script to skip downloading these files.
vi download_taxonomy.sh
########
Changed from
TAXONOMY_DIR="$KRAKEN2_DB_NAME/taxonomy"
if [ -z "$KRAKEN2_PROTEIN_DB" ]
To
for subsection in gb wgs
if [ -z "Viral" ]
########
sh download_taxonomy.sh
Prepare the genomes you’d like to add to your kraken database
You will have to look up the taxonomy ID and add this to each fasta headers as I have done below. i.e. (>sequence”|kraken:taxid|390850)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
#this just adds the taxids to the fasta headers, but does not affect sequence
bioawk -c fastx '{print ">"$name"|kraken:taxid|6326\n"$seq}' B.xylophilus.fa >B.xylophilusTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|166010\n"$seq}' D.destructor.fa >D.destructorTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|166011\n"$seq}' D.dipsaci.fa >D.dipsaciTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|1517492\n"$seq}' G.ellingtonae.fa >G.ellingtonaeTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|36090\n"$seq}' G.pallida.fa >G.pallidaTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|31243\n"$seq}' G.rostochiensis.fa >G.rostochiensisTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|6304\n"$seq}' M.arenaria.fa >M.arenariaTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|390850\n"$seq}' M.enterolobii.fa >M.enterolobiiTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|298850\n"$seq}' M.floridensis.fa >M.floridensisTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|189291\n"$seq}' M.graminicola.fa >M.graminicolaTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|6305\n"$seq}' M.hapla.fa >M.haplaTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|6306\n"$seq}' M.incognita.fa >M.incognitaTax.fa
bioawk -c fastx '{print ">"$name"|kraken:taxid|6303\n"$seq}' M.javanica.fa >M.javanicaTax.fa
bioawk -c fastx '{print ">"$name"kraken:taxid|51029\n"$seq}' H.glycines.fa >H.glycinesTax.fa
We are now ready to set the database to download and build
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
module use /work/GIF/software/modules
module load GIF/kraken2
module load blast-plus
kraken2-build --download-library viral --db NematodeViral
kraken2-build --download-library bacteria --db NematodeViral
kraken2-build --add-to-library B.xylophilusTax.fa -db NematodeViral
kraken2-build --add-to-library D.destructorTax.fa -db NematodeViral
kraken2-build --add-to-library D.dipsaciTax.fa -db NematodeViral
kraken2-build --add-to-library G.ellingtonaeTax.fa -db NematodeViral
kraken2-build --add-to-library G.pallidaTax.fa -db NematodeViral
kraken2-build --add-to-library G.rostochiensisTax.fa -db NematodeViral
kraken2-build --add-to-library H.glycinesTax.fa -db NematodeViral
kraken2-build --add-to-library M.arenariaTax.fa -db NematodeViral
kraken2-build --add-to-library M.enterolobiiTax.fa -db NematodeViral
kraken2-build --add-to-library M.floridensisTax.fa -db NematodeViral
kraken2-build --add-to-library M.graminicolaTax.fa -db NematodeViral
kraken2-build --add-to-library M.haplaTax.fa -db NematodeViral
kraken2-build --add-to-library M.incognitaTax.fa -db NematodeViral
kraken2-build --add-to-library M.javanicaTax.fa -db NematodeViral
kraken2-build --build --db NematodeViral --threads 16
#this took 5:26:32 on an hpc with 16 threads and 128GB ram.
Note that nematode genomes are small, this takes much longer for larger genomes.
Download RNASEQ samples that you’d like to classify with kraken
1
2
3
4
5
6
7
8
9
10
11
12
13
module load sra-toolkit
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files SRR3162514 #Globodera ellingtonae
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files SRR7775195 #Globodera pallida
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files ERR202492 #Globodera pallida
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files ERR202487 #Globodera rostochiensis
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files SRR7943144 #Ditylenchus destructor
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files DRR141214 #Bursaphelenchus xylophilus
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files ERR790020 # Meloidogyne javanica
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files SRR8691582 #Meloidogyne incognita
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files SRR2389452 #Meloidogyne graminicola
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files ERR790021 #Meloidogyne arenaria
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files SRR6269845 #Heterodera glycines
fastq-dump --outdir 04_DownloadedRNAseq/ --gzip --split-files SRR6269844 #Heterodera glycines
Create Kraken scripts
1
2
3
4
#note that you need to have the "#" in the output fastq files or kraken will crash.
paste <(ls -1 05_Reads/*_1*gz) <(ls -1 05_Reads/*_2*gz) |while read line; do echo "kraken2 -db NematodeViral --threads 16 --report "$line".report --gzip-compressed --unclassified-out "${line%.*}"unclassified#.fq --classified-out "${line%.*}"classified#.fq --paired "$line" > "${line%.*} ;done |awk '{print $1,$2,$3,$4,$5,$6,$8,$9,$10,$12,$13,$15,$16,$17,$18,$19,$21".Kraken.out"}' >kraken.sh
kraken.sh content
``` kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/DRR141214_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/DRR141214_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/DRR141214_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/DRR141214_1.fastq.gz 04_DownloadedRNAseq/DRR141214_2.fastq.gz > 04_DownloadedRNAseq/DRR141214_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/ERR202487_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/ERR202487_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/ERR202487_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/ERR202487_1.fastq.gz 04_DownloadedRNAseq/ERR202487_2.fastq.gz > 04_DownloadedRNAseq/ERR202487_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/ERR202492_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/ERR202492_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/ERR202492_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/ERR202492_1.fastq.gz 04_DownloadedRNAseq/ERR202492_2.fastq.gz > 04_DownloadedRNAseq/ERR202492_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/ERR790020_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/ERR790020_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/ERR790020_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/ERR790020_1.fastq.gz 04_DownloadedRNAseq/ERR790020_2.fastq.gz > 04_DownloadedRNAseq/ERR790020_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/SRR3162514_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/SRR3162514_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/SRR3162514_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/SRR3162514_1.fastq.gz 04_DownloadedRNAseq/SRR3162514_2.fastq.gz > 04_DownloadedRNAseq/SRR3162514_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/SRR7775195_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/SRR7775195_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/SRR7775195_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/SRR7775195_1.fastq.gz 04_DownloadedRNAseq/SRR7775195_2.fastq.gz > 04_DownloadedRNAseq/SRR7775195_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/SRR7943144_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/SRR7943144_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/SRR7943144_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/SRR7943144_1.fastq.gz 04_DownloadedRNAseq/SRR7943144_2.fastq.gz > 04_DownloadedRNAseq/SRR7943144_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/SRR7943144_2_val_2.fq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/SRR7943144_2_val_2.fqunclassified#.fq --classified-out 04_DownloadedRNAseq/SRR7943144_2_val_2.fqclassified#.fq --paired 04_DownloadedRNAseq/SRR7943144_1_val_1.fq.gz 04_DownloadedRNAseq/SRR7943144_2_val_2.fq.gz > 04_DownloadedRNAseq/SRR7943144_2_val_2.fq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/DRR141214_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/DRR141214_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/DRR141214_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/DRR141214_1.fastq.gz 04_DownloadedRNAseq/DRR141214_2.fastq.gz > 04_DownloadedRNAseq/DRR141214_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/ERR202487_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/ERR202487_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/ERR202487_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/ERR202487_1.fastq.gz 04_DownloadedRNAseq/ERR202487_2.fastq.gz > 04_DownloadedRNAseq/ERR202487_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/ERR202492_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/ERR202492_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/ERR202492_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/ERR202492_1.fastq.gz 04_DownloadedRNAseq/ERR202492_2.fastq.gz > 04_DownloadedRNAseq/ERR202492_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/ERR790020_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/ERR790020_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/ERR790020_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/ERR790020_1.fastq.gz 04_DownloadedRNAseq/ERR790020_2.fastq.gz > 04_DownloadedRNAseq/ERR790020_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/SRR3162514_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/SRR3162514_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/SRR3162514_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/SRR3162514_1.fastq.gz 04_DownloadedRNAseq/SRR3162514_2.fastq.gz > 04_DownloadedRNAseq/SRR3162514_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/SRR7775195_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/SRR7775195_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/SRR7775195_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/SRR7775195_1.fastq.gz 04_DownloadedRNAseq/SRR7775195_2.fastq.gz > 04_DownloadedRNAseq/SRR7775195_2.fastq.Kraken.out kraken2 -db NematodeViral --threads 16 --report 04_DownloadedRNAseq/SRR7943144_2.fastq.gz.report --gzip-compressed --unclassified-out 04_DownloadedRNAseq/SRR7943144_2.fastqunclassified#.fq --classified-out 04_DownloadedRNAseq/SRR7943144_2.fastqclassified#.fq --paired 04_DownloadedRNAseq/SRR7943144_1.fastq.gz 04_DownloadedRNAseq/SRR7943144_2.fastq.gz > 04_DownloadedRNAseq/SRR7943144_2.fastq.Kraken.out etc... ```
load the proper modules and submit to hpc
1
2
module load GIF/kraken2
module load perl
Summarize kraken data output
1
2
3
4
5
6
7
8
9
#generates list of only those with .01% of reads and removed nematodes from the report
for f in *report; do echo "awk '\$1>0 && \$3>100' "$f" |uniq|sort -k1,1nr |grep -v \"Meloidogyne\" |grep -v \"Heterodera\" |grep -v \"Globodera\" |grep -v \"Bursaphelenchus\" |grep -v \"Ditylenchus\" >"$f".summary" ;done >summarizer.sh
sh summarizer.sh
#example output from the script above.
#################################
awk '$1>0 && $3>100' Ga1-pol-1_S1_L004_R1_001.fastq.gz.report |uniq|sort -k1,1nr|grep -v "Meloidogyne" |grep -v "Heterodera" |grep -v "Globodera" |grep -v "Bursaphelenchus" |grep -v "Ditylenchus" >Ga1-pol-1_S1_L004_R1_001.fastq.gz.report.summary
#################################
I took these files, added the species name to the fifth column, removed those entries that had fewer than 100 reads allocated, kept only genera, species, and subspecies, and then concatenated all files for a network in cytoscape.
Summarized output from all samples
Click to see the results of each run below.
Bursaphelenchus xylophilus DRR1414214 "pine wilt nematode"
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|---------|---------------------------------|-------------------|---------------| | 99.18 | 23932404 | 831 | O2 | 33283 | Tylenchomorpha | B. xylophilus | DRR1414214 | | 0.69 | 166444 | 166444 | U | 0 | unclassified | B. xylophilus | DRR1414214 | | 0.13 | 30825 | 357 | D | 2 | Bacteria | B. xylophilus | DRR1414214 | | 0.08 | 18597 | 235 | D1 | 1783272 | Terrabacteria group | B. xylophilus | DRR1414214 | | 0.07 | 16925 | 16925 | S | 1286 | Staphylococcus simulans | B. xylophilus | DRR1414214 | | 0.07 | 16992 | 30 | G | 1279 | Staphylococcus | B. xylophilus | DRR1414214 | | 0.05 | 11743 | 519 | P | 1224 | Proteobacteria | B. xylophilus | DRR1414214 | | 0.04 | 9035 | 446 | C | 1236 | Gammaproteobacteria | B. xylophilus | DRR1414214 | | 0.03 | 7195 | 7195 | S2 | 1365647 | Xanthomonas euvesicatoria pv. alfalfae CFBP 3836 | B. xylophilus | DRR1414214 |
Globodera rostochiensis ERR202487 "yellow potato cyst nematode"
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|---------|---------------------|------------------|-----------| | 98.46 | 28153971 | 832 | O2 | 33283 | Tylenchomorpha | G. rostochiensis | ERR202487 | | 98.45 | 28151761 | 5340 | O3 | 33284 | Tylenchoidea | G. rostochiensis | ERR202487 | | 98.41 | 28139651 | 173770 | F1 | 33286 | Heteroderinae | G. rostochiensis | ERR202487 | | 1.5 | 429425 | 429425 | U | 0 | unclassified | G. rostochiensis | ERR202487 | | 0.03 | 9513 | 516 | D | 2 | Bacteria | G. rostochiensis | ERR202487 | | 0.01 | 1748 | 38 | C | 1236 | Gammaproteobacteria | G. rostochiensis | ERR202487 | | 0.01 | 3266 | 12 | D1 | 1783272 | Terrabacteria group | G. rostochiensis | ERR202487 | | 0.01 | 4116 | 151 | P | 1224 | Proteobacteria | G. rostochiensis | ERR202487 |
Globodera pallida ERR202492 "white potato cyst nematode"
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|-------|---------------------|----------------|-----------| | 99.22 | 30582562 | 2151 | O3 | 33284 | Tylenchoidea | G. pallida | ERR202492 | | 99.22 | 30583755 | 507 | O2 | 33283 | Tylenchomorpha | G. pallida | ERR202492 | | 99.21 | 30578964 | 71917 | F1 | 33286 | Heteroderinae | G. pallida | ERR202492 | | 0.74 | 228813 | 228813 | U | 0 | unclassified | G. pallida | ERR202492 | | 0.03 | 9354 | 252 | D | 2 | Bacteria | G. pallida | ERR202492 | | 0.02 | 5572 | 282 | P | 1224 | Proteobacteria | G. pallida | ERR202492 | | 0.01 | 1836 | 402 | F | 75682 | Oxalobacteraceae | G. pallida | ERR202492 | | 0.01 | 1942 | 115 | C | 1236 | Gammaproteobacteria | G. pallida | ERR202492 | | 0.01 | 2323 | 155 | O | 80840 | Burkholderiales | G. pallida | ERR202492 | | 0.01 | 2458 | 86 | C | 28216 | Betaproteobacteria | G. pallida | ERR202492 |
Meloidogyne javanica ERR790020 "sugarcane eelworm"
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|---------|------------------------------------------------|----------------|-----------| | 85.23 | 26295871 | 631 | O2 | 33283 | Tylenchomorpha | M. javanica | ERR790020 | | 85.18 | 26278729 | 985 | O3 | 33284 | Tylenchoidea | M. javanica | ERR790020 | | 14.28 | 4404990 | 4404990 | U | 0 | unclassified | M. javanica | ERR790020 | | 0.45 | 138563 | 1002 | D | 2 | Bacteria | M. javanica | ERR790020 | | 0.26 | 78919 | 125 | D1 | 1783272 | Terrabacteria group | M. javanica | ERR790020 | | 0.21 | 63936 | 55 | P | 1239 | Firmicutes | M. javanica | ERR790020 | | 0.14 | 43539 | 34 | F | 186817 | Bacillaceae | M. javanica | ERR790020 | | 0.13 | 39433 | 488 | P | 1224 | Proteobacteria | M. javanica | ERR790020 | | 0.12 | 36797 | 35836 | G1 | 86661 | Bacillus cereus group | M. javanica | ERR790020 | | 0.12 | 37775 | 142 | G | 1386 | Bacillus | M. javanica | ERR790020 | | 0.09 | 27321 | 122 | C | 1236 | Gammaproteobacteria | M. javanica | ERR790020 | | 0.04 | 10990 | 83 | C | 1760 | Actinobacteria | M. javanica | ERR790020 | | 0.04 | 11634 | 11634 | S | 91844 | Candidatus Portiera aleyrodidarum | M. javanica | ERR790020 | | 0.03 | 8041 | 65 | F1 | 33286 | Heteroderinae | M. javanica | ERR790020 | | 0.02 | 4727 | 24 | O | 91347 | Enterobacterales | M. javanica | ERR790020 | | 0.02 | 5469 | 5469 | S | 1570 | Halobacillus halophilus | M. javanica | ERR790020 | | 0.02 | 5554 | 22 | F | 49546 | Flavobacteriaceae | M. javanica | ERR790020 | | 0.02 | 5763 | 5763 | S | 51515 | Dehalobacterium formicoaceticum | M. javanica | ERR790020 | | 0.02 | 6065 | 67 | G | 1485 | Clostridium | M. javanica | ERR790020 | | 0.02 | 7553 | 7553 | S1 | 331113 | Simkania negevensis Z | M. javanica | ERR790020 | | 0.01 | 1851 | 22 | G | 237 | Flavobacterium | M. javanica | ERR790020 | | 0.01 | 1903 | 22 | F | 85023 | Microbacteriaceae | M. javanica | ERR790020 | | 0.01 | 1994 | 16 | P | 1117 | Cyanobacteria | M. javanica | ERR790020 | | 0.01 | 2120 | 218 | G | 286 | Pseudomonas | M. javanica | ERR790020 | | 0.01 | 2191 | 2171 | S | 1747 | Cutibacterium acnes | M. javanica | ERR790020 | | 0.01 | 2198 | 2198 | S | 12253 | Tomato mosaic virus | M. javanica | ERR790020 | | 0.01 | 2278 | 24 | G | 1912216 | Cutibacterium | M. javanica | ERR790020 | | 0.01 | 2348 | 26 | F | 31957 | Propionibacteriaceae | M. javanica | ERR790020 | | 0.01 | 2405 | 2405 | S1 | 1874362 | Campylobacter pinnipediorum subsp. caledonicus | M. javanica | ERR790020 | | 0.01 | 2487 | 2487 | S | 646637 | Streptomyces sp. M2 | M. javanica | ERR790020 | | 0.01 | 2543 | 173 | G | 469 | Acinetobacter | M. javanica | ERR790020 | | 0.01 | 2550 | 397 | G | 1301 | Streptococcus | M. javanica | ERR790020 | | 0.01 | 2692 | 11 | G | 194 | Campylobacter | M. javanica | ERR790020 | | 0.01 | 2692 | 2692 | S | 1492 | Clostridium butyricum | M. javanica | ERR790020 | | 0.01 | 2840 | 11 | O | 80840 | Burkholderiales | M. javanica | ERR790020 | | 0.01 | 2901 | 15 | C | 28211 | Alphaproteobacteria | M. javanica | ERR790020 | | 0.01 | 3450 | 634 | F | 543 | Enterobacteriaceae | M. javanica | ERR790020 | | 0.01 | 3859 | 11 | O | 186826 | Lactobacillales | M. javanica | ERR790020 | | 0.01 | 4358 | 179 | G | 1883 | Streptomyces | M. javanica | ERR790020 |
Meloidogyne arenaria ERR790021 peanut root knot nematode
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|---------|------------------------------------------------|----------------|-----------| | 83.62 | 30208155 | 623 | O2 | 33283 | Tylenchomorpha | M. arenaria | ERR790021 | | 83.59 | 30200098 | 1194 | O3 | 33284 | Tylenchoidea | M. arenaria | ERR790021 | | 15.86 | 5729345 | 5729345 | U | 0 | unclassified | M. arenaria | ERR790021 | | 0.49 | 175784 | 796 | D | 2 | Bacteria | M. arenaria | ERR790021 | | 0.31 | 111238 | 154 | D1 | 1783272 | Terrabacteria group | M. arenaria | ERR790021 | | 0.22 | 78559 | 50 | P | 1239 | Firmicutes | M. arenaria | ERR790021 | | 0.18 | 65256 | 13 | C | 91061 | Bacilli | M. arenaria | ERR790021 | | 0.17 | 60002 | 11 | O | 1385 | Bacillales | M. arenaria | ERR790021 | | 0.15 | 53041 | 88 | F | 186817 | Bacillaceae | M. arenaria | ERR790021 | | 0.14 | 49114 | 187 | G | 1386 | Bacillus | M. arenaria | ERR790021 | | 0.13 | 47856 | 45296 | G1 | 86661 | Bacillus cereus group | M. arenaria | ERR790021 | | 0.1 | 35545 | 892 | P | 1224 | Proteobacteria | M. arenaria | ERR790021 | | 0.08 | 28005 | 91 | C | 1760 | Actinobacteria | M. arenaria | ERR790021 | | 0.05 | 16321 | 118 | F | 31957 | Propionibacteriaceae | M. arenaria | ERR790021 | | 0.04 | 14709 | 141 | C | 1236 | Gammaproteobacteria | M. arenaria | ERR790021 | | 0.04 | 15264 | 14533 | S | 1747 | Cutibacterium acnes | M. arenaria | ERR790021 | | 0.04 | 16033 | 104 | G | 1912216 | Cutibacterium | M. arenaria | ERR790021 | | 0.03 | 11344 | 74 | F1 | 33286 | Heteroderinae | M. arenaria | ERR790021 | | 0.02 | 5597 | 5597 | S | 2029983 | Chitinophaga caeni | M. arenaria | ERR790021 | | 0.02 | 5865 | 5865 | S1 | 331113 | Simkania negevensis Z | M. arenaria | ERR790021 | | 0.02 | 7082 | 283 | G | 1485 | Clostridium | M. arenaria | ERR790021 | | 0.02 | 7943 | 18 | F | 49546 | Flavobacteriaceae | M. arenaria | ERR790021 | | 0.02 | 8846 | 39 | C | 28211 | Alphaproteobacteria | M. arenaria | ERR790021 | | 0.01 | 1916 | 1916 | S | 1536774 | Paenibacillus sp. FSL H7-0357 | M. arenaria | ERR790021 | | 0.01 | 1935 | 184 | G | 286 | Pseudomonas | M. arenaria | ERR790021 | | 0.01 | 1963 | 16 | O | 85007 | Corynebacteriales | M. arenaria | ERR790021 | | 0.01 | 2029 | 18 | O | 135622 | Alteromonadales | M. arenaria | ERR790021 | | 0.01 | 2069 | 2069 | S | 1678728 | Flavobacterium kingsejongi | M. arenaria | ERR790021 | | 0.01 | 2280 | 15 | O | 80840 | Burkholderiales | M. arenaria | ERR790021 | | 0.01 | 2349 | 19 | G | 44249 | Paenibacillus | M. arenaria | ERR790021 | | 0.01 | 2568 | 2568 | S | 1492 | Clostridium butyricum | M. arenaria | ERR790021 | | 0.01 | 2618 | 51 | G | 237 | Flavobacterium | M. arenaria | ERR790021 | | 0.01 | 2973 | 2973 | S | 51515 | Dehalobacterium formicoaceticum | M. arenaria | ERR790021 | | 0.01 | 3041 | 455 | F | 543 | Enterobacteriaceae | M. arenaria | ERR790021 | | 0.01 | 3132 | 104 | G | 1279 | Staphylococcus | M. arenaria | ERR790021 | | 0.01 | 3340 | 3340 | S | 1570 | Halobacillus halophilus | M. arenaria | ERR790021 | | 0.01 | 3458 | 598 | G | 1301 | Streptococcus | M. arenaria | ERR790021 | | 0.01 | 3538 | 3538 | S | 2268461 | Microbacterium sp. ABRD_28 | M. arenaria | ERR790021 | | 0.01 | 3692 | 26 | G | 33882 | Microbacterium | M. arenaria | ERR790021 | | 0.01 | 3920 | 177 | G | 1883 | Streptomyces | M. arenaria | ERR790021 | | 0.01 | 4424 | 4424 | S | 2500532 | Paracoccus sp. Arc7-R13 | M. arenaria | ERR790021 | | 0.01 | 4699 | 4699 | S1 | 1874362 | Campylobacter pinnipediorum subsp. caledonicus | M. arenaria | ERR790021 | | 0.01 | 4746 | 29 | G | 265 | Paracoccus | M. arenaria | ERR790021 | | 0.01 | 5058 | 35 | O | 91347 | Enterobacterales | M. arenaria | ERR790021 | | 0.01 | 5197 | 29 | F | 31989 | Rhodobacteraceae | M. arenaria | ERR790021 | | 0.01 | 5360 | 19 | G | 194 | Campylobacter | M. arenaria | ERR790021 |
Globodera pallida SRR2389452 "white potato cyst nematode"
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|---------|-----------------------------------------------|----------------|------------| | 70.18 | 38275890 | 323730 | O2 | 33283 | Tylenchomorpha | G. pallida | SRR2389452 | | 69.28 | 37787560 | 542522 | O3 | 33284 | Tylenchoidea | G. pallida | SRR2389452 | | 28.39 | 15481907 | 15481907 | U | 0 | unclassified | G. pallida | SRR2389452 | | 1.42 | 774660 | 19941 | F1 | 33286 | Heteroderinae | G. pallida | SRR2389452 | | 1.17 | 636686 | 27777 | D | 2 | Bacteria | G. pallida | SRR2389452 | | 0.79 | 428302 | 20022 | P | 1224 | Proteobacteria | G. pallida | SRR2389452 | | 0.4 | 220524 | 34307 | C | 28216 | Betaproteobacteria | G. pallida | SRR2389452 | | 0.27 | 146239 | 22120 | O | 80840 | Burkholderiales | G. pallida | SRR2389452 | | 0.18 | 97552 | 8903 | C | 28211 | Alphaproteobacteria | G. pallida | SRR2389452 | | 0.16 | 87901 | 1832 | D1 | 1783272 | Terrabacteria group | G. pallida | SRR2389452 | | 0.15 | 80414 | 5058 | P | 976 | Bacteroidetes | G. pallida | SRR2389452 | | 0.15 | 81131 | 165 | D2 | 68336 | Bacteroidetes/Chlorobi group | G. pallida | SRR2389452 | | 0.15 | 81358 | 12 | D1 | 1783270 | FCB group | G. pallida | SRR2389452 | | 0.15 | 81832 | 7179 | C | 1236 | Gammaproteobacteria | G. pallida | SRR2389452 | | 0.1 | 54704 | 20943 | F | 75682 | Oxalobacteraceae | G. pallida | SRR2389452 | | 0.09 | 48210 | 652 | P | 1239 | Firmicutes | G. pallida | SRR2389452 | | 0.05 | 25469 | 987 | O | 204457 | Sphingomonadales | G. pallida | SRR2389452 | | 0.05 | 25978 | 2756 | F | 41295 | Rhodospirillaceae | G. pallida | SRR2389452 | | 0.05 | 28012 | 439 | C | 91061 | Bacilli | G. pallida | SRR2389452 | | 0.05 | 28082 | 279 | O | 72274 | Pseudomonadales | G. pallida | SRR2389452 | | 0.05 | 28199 | 827 | O | 204441 | Rhodospirillales | G. pallida | SRR2389452 | | 0.05 | 29673 | 10207 | F | 80864 | Comamonadaceae | G. pallida | SRR2389452 | | 0.04 | 20115 | 20115 | S | 1922225 | Erythrobacter sp. Alg231-14 | G. pallida | SRR2389452 | | 0.04 | 20368 | 54 | G | 1041 | Erythrobacter | G. pallida | SRR2389452 | | 0.04 | 20590 | 570 | O | 1385 | Bacillales | G. pallida | SRR2389452 | | 0.04 | 20893 | 5580 | F | 119060 | Burkholderiaceae | G. pallida | SRR2389452 | | 0.04 | 20936 | 187 | F | 335929 | Erythrobacteraceae | G. pallida | SRR2389452 | | 0.04 | 21140 | 4897 | F | 49546 | Flavobacteriaceae | G. pallida | SRR2389452 | | 0.04 | 22234 | 456 | O | 200644 | Flavobacteriales | G. pallida | SRR2389452 | | 0.04 | 22653 | 3727 | O | 206351 | Neisseriales | G. pallida | SRR2389452 | | 0.04 | 23270 | 1926 | C | 1760 | Actinobacteria | G. pallida | SRR2389452 | | 0.04 | 24528 | 204 | P | 201174 | Actinobacteria | G. pallida | SRR2389452 | | 0.03 | 13690 | 2029 | F | 76892 | Caulobacteraceae | G. pallida | SRR2389452 | | 0.03 | 13981 | 215 | F | 186817 | Bacillaceae | G. pallida | SRR2389452 | | 0.03 | 14763 | 2006 | O | 356 | Rhizobiales | G. pallida | SRR2389452 | | 0.03 | 15303 | 1910 | O | 768507 | Cytophagales | G. pallida | SRR2389452 | | 0.03 | 15341 | 8994 | G | 469 | Acinetobacter | G. pallida | SRR2389452 | | 0.03 | 16420 | 3898 | F | 563835 | Chitinophagaceae | G. pallida | SRR2389452 | | 0.03 | 16735 | 188 | O | 186802 | Clostridiales | G. pallida | SRR2389452 | | 0.03 | 16854 | 5100 | F | 1499392 | Chromobacteriaceae | G. pallida | SRR2389452 | | 0.03 | 16899 | 2661 | F | 84566 | Sphingobacteriaceae | G. pallida | SRR2389452 | | 0.03 | 17540 | 77 | C | 186801 | Clostridia | G. pallida | SRR2389452 | | 0.03 | 17545 | 1221 | F | 468 | Moraxellaceae | G. pallida | SRR2389452 | | 0.03 | 18205 | 13606 | G | 191 | Azospirillum | G. pallida | SRR2389452 | | 0.03 | 18569 | 8142 | G | 149698 | Massilia | G. pallida | SRR2389452 | | 0.02 | 10258 | 574 | F | 135621 | Pseudomonadaceae | G. pallida | SRR2389452 | | 0.02 | 10859 | 1220 | O | 206389 | Rhodocyclales | G. pallida | SRR2389452 | | 0.02 | 11013 | 5039 | O2 | 224471 | Burkholderiales Genera incertae sedis | G. pallida | SRR2389452 | | 0.02 | 11580 | 1633 | G | 1386 | Bacillus | G. pallida | SRR2389452 | | 0.02 | 11637 | 7961 | G | 963 | Herbaspirillum | G. pallida | SRR2389452 | | 0.02 | 13397 | 1273 | O | 91347 | Enterobacterales | G. pallida | SRR2389452 | | 0.02 | 13575 | 1982 | O1 | 119065 | unclassified Burkholderiales | G. pallida | SRR2389452 | | 0.02 | 8205 | 2436 | G | 79328 | Chitinophaga | G. pallida | SRR2389452 | | 0.02 | 8275 | 2424 | F | 543 | Enterobacteriaceae | G. pallida | SRR2389452 | | 0.02 | 8344 | 247 | G | 47420 | Hydrogenophaga | G. pallida | SRR2389452 | | 0.02 | 8960 | 292 | F | 89373 | Cytophagaceae | G. pallida | SRR2389452 | | 0.02 | 9593 | 2478 | G | 286 | Pseudomonas | G. pallida | SRR2389452 | | 0.01 | 2739 | 2739 | S | 29442 | Pseudomonas tolaasii | G. pallida | SRR2389452 | | 0.01 | 2860 | 1306 | G | 535 | Chromobacterium | G. pallida | SRR2389452 | | 0.01 | 2927 | 2414 | G | 75 | Caulobacter | G. pallida | SRR2389452 | | 0.01 | 3144 | 11 | O | 213849 | Campylobacterales | G. pallida | SRR2389452 | | 0.01 | 3159 | 578 | F | 31989 | Rhodobacteraceae | G. pallida | SRR2389452 | | 0.01 | 3177 | 3153 | S | 48296 | Acinetobacter pittii | G. pallida | SRR2389452 | | 0.01 | 3191 | 11 | C | 29547 | Epsilonproteobacteria | G. pallida | SRR2389452 | | 0.01 | 3210 | 3210 | S | 1678028 | Massilia sp. NR 4-1 | G. pallida | SRR2389452 | | 0.01 | 3282 | 48 | G1 | 136843 | Pseudomonas fluorescens group | G. pallida | SRR2389452 | | 0.01 | 3315 | 292 | O | 171549 | Bacteroidales | G. pallida | SRR2389452 | | 0.01 | 3324 | 773 | F1 | 227290 | Rhizobium/Agrobacterium group | G. pallida | SRR2389452 | | 0.01 | 3336 | 830 | F | 85023 | Microbacteriaceae | G. pallida | SRR2389452 | | 0.01 | 3496 | 13 | O | 204455 | Rhodobacterales | G. pallida | SRR2389452 | | 0.01 | 3546 | 676 | F | 41297 | Sphingomonadaceae | G. pallida | SRR2389452 | | 0.01 | 3857 | 29 | G1 | 909768 | Acinetobacter calcoaceticus/baumannii complex | G. pallida | SRR2389452 | | 0.01 | 3858 | 54 | C | 31969 | Mollicutes | G. pallida | SRR2389452 | | 0.01 | 3916 | 22 | C | 200643 | Bacteroidia | G. pallida | SRR2389452 | | 0.01 | 3973 | 2523 | S | 1491 | Clostridium botulinum | G. pallida | SRR2389452 | | 0.01 | 4050 | 859 | G | 1883 | Streptomyces | G. pallida | SRR2389452 | | 0.01 | 4127 | 198 | O | 85007 | Corynebacteriales | G. pallida | SRR2389452 | | 0.01 | 4207 | 205 | C | 28221 | Deltaproteobacteria | G. pallida | SRR2389452 | | 0.01 | 4317 | 89 | F | 2062 | Streptomycetaceae | G. pallida | SRR2389452 | | 0.01 | 4447 | 88 | O | 135622 | Alteromonadales | G. pallida | SRR2389452 | | 0.01 | 4482 | 468 | F | 32033 | Xanthomonadaceae | G. pallida | SRR2389452 | | 0.01 | 4513 | 491 | F | 82115 | Rhizobiaceae | G. pallida | SRR2389452 | | 0.01 | 4549 | 1675 | G | 237 | Flavobacterium | G. pallida | SRR2389452 | | 0.01 | 4708 | 4708 | S1 | 485918 | Chitinophaga pinensis DSM 2588 | G. pallida | SRR2389452 | | 0.01 | 4908 | 615 | F | 506 | Alcaligenaceae | G. pallida | SRR2389452 | | 0.01 | 5028 | 3677 | G | 28453 | Sphingobacterium | G. pallida | SRR2389452 | | 0.01 | 5058 | 5058 | S1 | 526973 | Bacillus cereus m1293 | G. pallida | SRR2389452 | | 0.01 | 5225 | 245 | O | 32003 | Nitrosomonadales | G. pallida | SRR2389452 | | 0.01 | 5504 | 5504 | S | 2306583 | Halomonas sp. JS92-SW72 | G. pallida | SRR2389452 | | 0.01 | 5559 | 5559 | S | 1677857 | Hungateiclostridium saccincola | G. pallida | SRR2389452 | | 0.01 | 5705 | 3302 | G | 59732 | Chryseobacterium | G. pallida | SRR2389452 | | 0.01 | 5826 | 5826 | S | 748280 | Pseudogulbenkiania sp. NH8B | G. pallida | SRR2389452 | | 0.01 | 5860 | 269 | O | 135614 | Xanthomonadales | G. pallida | SRR2389452 | | 0.01 | 6030 | 3679 | G | 73029 | Dechloromonas | G. pallida | SRR2389452 | | 0.01 | 6163 | 349 | S | 1396 | Bacillus cereus | G. pallida | SRR2389452 | | 0.01 | 6184 | 2141 | G | 32008 | Burkholderia | G. pallida | SRR2389452 | | 0.01 | 6481 | 715 | G | 2745 | Halomonas | G. pallida | SRR2389452 | | 0.01 | 6785 | 40 | F | 28256 | Halomonadaceae | G. pallida | SRR2389452 | | 0.01 | 6983 | 510 | O | 186826 | Lactobacillales | G. pallida | SRR2389452 | | 0.01 | 7050 | 305 | G1 | 86661 | Bacillus cereus group | G. pallida | SRR2389452 | | 0.01 | 7260 | 1093 | P | 1117 | Cyanobacteria | G. pallida | SRR2389452 | | 0.01 | 7272 | 1510 | O | 85006 | Micrococcales | G. pallida | SRR2389452 | | 0.01 | 7285 | 2691 | G | 423349 | Mucilaginibacter | G. pallida | SRR2389452 | | 0.01 | 7572 | 66 | O | 135619 | Oceanospirillales | G. pallida | SRR2389452 | | 0.01 | 7650 | 7512 | S | 78587 | Asticcacaulis excentricus | G. pallida | SRR2389452 | | 0.01 | 7772 | 7772 | S | 2184519 | Hydrogenophaga sp. NH-16 | G. pallida | SRR2389452 | | 0.01 | 7823 | 678 | G | 1485 | Clostridium | G. pallida | SRR2389452 | | 0.01 | 8050 | 25 | F | 31979 | Clostridiaceae | G. pallida | SRR2389452 |
Globodera ellingtonae SRR3162514 "potato cyst nematode"
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|---------|--------------------------------------------------|----------------|------------| | 93.97 | 28073787 | 1899 | O2 | 33283 | Tylenchomorpha | G. ellingtonae | SRR3162514 | | 93.95 | 28070030 | 7449 | O3 | 33284 | Tylenchoidea | G. ellingtonae | SRR3162514 | | 93.91 | 28057567 | 40650 | F1 | 33286 | Heteroderinae | G. ellingtonae | SRR3162514 | | 4.99 | 1491074 | 1491074 | U | 0 | unclassified | G. ellingtonae | SRR3162514 | | 0.13 | 39802 | 1626 | D | 2 | Bacteria | G. ellingtonae | SRR3162514 | | 0.1 | 28497 | 20210 | P | 1224 | Proteobacteria | G. ellingtonae | SRR3162514 | | 0.03 | 7472 | 30 | D1 | 1783272 | Terrabacteria group | G. ellingtonae | SRR3162514 | | 0.02 | 4515 | 103 | C | 1236 | Gammaproteobacteria | G. ellingtonae | SRR3162514 | | 0.01 | 1708 | 30 | P | 976 | Bacteroidetes | G. ellingtonae | SRR3162514 | | 0.01 | 1825 | 1825 | S2 | 1365647 | Xanthomonas euvesicatoria pv. alfalfae CFBP 3836 | G. ellingtonae | SRR3162514 | | 0.01 | 1915 | 22 | O | 1385 | Bacillales | G. ellingtonae | SRR3162514 | | 0.01 | 1954 | 1954 | S1 | 1200984 | Streptomyces lividans 1326 | G. ellingtonae | SRR3162514 | | 0.01 | 1989 | 60 | C | 28211 | Alphaproteobacteria | G. ellingtonae | SRR3162514 | | 0.01 | 2194 | 72 | G | 1883 | Streptomyces | G. ellingtonae | SRR3162514 | | 0.01 | 2338 | 11 | C | 91061 | Bacilli | G. ellingtonae | SRR3162514 | | 0.01 | 3255 | 18 | P | 1239 | Firmicutes | G. ellingtonae | SRR3162514 | | 0.01 | 3462 | 90 | C | 1760 | Actinobacteria | G. ellingtonae | SRR3162514 |
Heterodera glycines SRR6269844 "soybean cyst nematode"
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|---------|---------------------------------------|----------------|------------| | 93.3 | 10730672 | 79467 | O2 | 33283 | Tylenchomorpha | H. glycines | SRR6269844 | | 92.54 | 10642968 | 66339 | O3 | 33284 | Tylenchoidea | H. glycines | SRR6269844 | | 90.73 | 10434857 | 1014172 | F1 | 33286 | Heteroderinae | H. glycines | SRR6269844 | | 5.26 | 605129 | 341864 | D | 2 | Bacteria | H. glycines | SRR6269844 | | 1.28 | 147037 | 147037 | U | 0 | unclassified | H. glycines | SRR6269844 | | 1.15 | 131998 | 64497 | P | 1224 | Proteobacteria | H. glycines | SRR6269844 | | 1.09 | 125245 | 67 | D1 | 1783272 | Terrabacteria group | H. glycines | SRR6269844 | | 1.08 | 123897 | 41 | P | 1239 | Firmicutes | H. glycines | SRR6269844 | | 1.07 | 122930 | 17 | O | 1385 | Bacillales | H. glycines | SRR6269844 | | 1.07 | 123180 | 11 | C | 91061 | Bacilli | H. glycines | SRR6269844 | | 0.81 | 93587 | 2453 | G | 1279 | Staphylococcus | H. glycines | SRR6269844 | | 0.79 | 90934 | 90934 | S | 1286 | Staphylococcus simulans | H. glycines | SRR6269844 | | 0.35 | 40388 | 779 | C | 28216 | Betaproteobacteria | H. glycines | SRR6269844 | | 0.34 | 39339 | 1612 | O | 80840 | Burkholderiales | H. glycines | SRR6269844 | | 0.24 | 27634 | 27634 | S1 | 526973 | Bacillus cereus m1293 | H. glycines | SRR6269844 | | 0.24 | 27679 | 36 | S | 1396 | Bacillus cereus | H. glycines | SRR6269844 | | 0.24 | 27741 | 29 | G1 | 86661 | Bacillus cereus group | H. glycines | SRR6269844 | | 0.24 | 27987 | 34 | G | 1386 | Bacillus | H. glycines | SRR6269844 | | 0.16 | 18542 | 80 | C | 1236 | Gammaproteobacteria | H. glycines | SRR6269844 | | 0.16 | 18658 | 1587 | F | 80864 | Comamonadaceae | H. glycines | SRR6269844 | | 0.14 | 15602 | 15602 | S | 2306583 | Halomonas sp. JS92-SW72 | H. glycines | SRR6269844 | | 0.12 | 14279 | 14279 | S | 2184519 | Hydrogenophaga sp. NH-16 | H. glycines | SRR6269844 | | 0.1 | 11137 | 859 | O1 | 119065 | unclassified Burkholderiales | H. glycines | SRR6269844 | | 0.08 | 9457 | 3902 | O2 | 224471 | Burkholderiales Genera incertae sedis | H. glycines | SRR6269844 | | 0.07 | 8145 | 182 | C | 28211 | Alphaproteobacteria | H. glycines | SRR6269844 | | 0.05 | 5199 | 27 | P | 976 | Bacteroidetes | H. glycines | SRR6269844 | | 0.05 | 5207 | 836 | F | 119060 | Burkholderiaceae | H. glycines | SRR6269844 | | 0.05 | 6321 | 6321 | S | 1034377 | Soybean cyst nematode socyvirus | H. glycines | SRR6269844 | | 0.03 | 3516 | 183 | G | 107 | Spirosoma | H. glycines | SRR6269844 | | 0.03 | 3678 | 17 | O | 768507 | Cytophagales | H. glycines | SRR6269844 | | 0.02 | 1853 | 1846 | G1 | 114292 | typhus group | H. glycines | SRR6269844 | | 0.02 | 1897 | 12 | G | 780 | Rickettsia | H. glycines | SRR6269844 | | 0.02 | 2124 | 2124 | S | 2057025 | Spirosoma pollinicola | H. glycines | SRR6269844 | | 0.02 | 2281 | 2281 | S1 | 395495 | Leptothrix cholodnii SP-6 | H. glycines | SRR6269844 | | 0.02 | 2408 | 2408 | S | 96344 | Cupriavidus oxalaticus | H. glycines | SRR6269844 | | 0.02 | 2586 | 312 | F | 41297 | Sphingomonadaceae | H. glycines | SRR6269844 | | 0.02 | 2592 | 641 | F | 75682 | Oxalobacteraceae | H. glycines | SRR6269844 | | 0.02 | 2873 | 226 | O | 204457 | Sphingomonadales | H. glycines | SRR6269844 | | 0.01 | 1002 | 262 | G | 44249 | Paenibacillus | H. glycines | SRR6269844 | | 0.01 | 1051 | 179 | G | 149698 | Massilia | H. glycines | SRR6269844 | | 0.01 | 1076 | 86 | F | 76892 | Caulobacteraceae | H. glycines | SRR6269844 | | 0.01 | 1191 | 80 | F | 186822 | Paenibacillaceae | H. glycines | SRR6269844 | | 0.01 | 1199 | 622 | G | 13687 | Sphingomonas | H. glycines | SRR6269844 | | 0.01 | 1255 | 95 | O | 356 | Rhizobiales | H. glycines | SRR6269844 | | 0.01 | 1332 | 53 | G | 286 | Pseudomonas | H. glycines | SRR6269844 | | 0.01 | 1631 | 1250 | G | 48736 | Ralstonia | H. glycines | SRR6269844 | | 0.01 | 1690 | 1690 | S | 1768242 | Paucibacter sp. KCTC 42545 | H. glycines | SRR6269844 | | 0.01 | 587 | 557 | G | 165695 | Sphingobium | H. glycines | SRR6269844 | | 0.01 | 669 | 122 | G | 20 | Phenylobacterium | H. glycines | SRR6269844 | | 0.01 | 684 | 684 | S | 1211326 | Spirosoma aerolatum | H. glycines | SRR6269844 | | 0.01 | 766 | 503 | G | 963 | Herbaspirillum | H. glycines | SRR6269844 | | 0.01 | 800 | 800 | S | 864051 | Burkholderiales bacterium JOSHI_001 | H. glycines | SRR6269844 | | 0.01 | 857 | 111 | G | 283 | Comamonas | H. glycines | SRR6269844 | | 0.01 | 881 | 881 | S1 | 390235 | Pseudomonas putida W619 | H. glycines | SRR6269844 |
Heterodera glycines SRR6269845 "soybean cyst nematode"
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|---------|---------------------------------------|----------------|------------| | 92.88 | 11978072 | 122706 | O2 | 33283 | Tylenchomorpha | H. glycines | SRR6269845 | | 91.8 | 11838666 | 96582 | O3 | 33284 | Tylenchoidea | H. glycines | SRR6269845 | | 90.32 | 11648223 | 1223381 | F1 | 33286 | Heteroderinae | H. glycines | SRR6269845 | | 6.04 | 778844 | 4308 | D | 2 | Bacteria | H. glycines | SRR6269845 | | 5.84 | 753772 | 6723 | P | 1224 | Proteobacteria | H. glycines | SRR6269845 | | 5.66 | 730524 | 1453 | C | 28216 | Betaproteobacteria | H. glycines | SRR6269845 | | 5.65 | 728391 | 1552 | O | 80840 | Burkholderiales | H. glycines | SRR6269845 | | 5.26 | 678448 | 267 | F | 119060 | Burkholderiaceae | H. glycines | SRR6269845 | | 5.25 | 677415 | 677415 | S | 96344 | Cupriavidus oxalaticus | H. glycines | SRR6269845 | | 5.25 | 677466 | 16 | G | 106589 | Cupriavidus | H. glycines | SRR6269845 | | 0.97 | 124856 | 124856 | U | 0 | unclassified | H. glycines | SRR6269845 | | 0.21 | 26534 | 1789 | F | 80864 | Comamonadaceae | H. glycines | SRR6269845 | | 0.17 | 22023 | 22023 | S | 2184519 | Hydrogenophaga sp. NH-16 | H. glycines | SRR6269845 | | 0.15 | 18854 | 2275 | O1 | 119065 | unclassified Burkholderiales | H. glycines | SRR6269845 | | 0.13 | 16283 | 37 | D1 | 1783272 | Terrabacteria group | H. glycines | SRR6269845 | | 0.12 | 15206 | 14 | P | 1239 | Firmicutes | H. glycines | SRR6269845 | | 0.12 | 15729 | 6855 | O2 | 224471 | Burkholderiales Genera incertae sedis | H. glycines | SRR6269845 | | 0.11 | 14431 | 11 | O | 1385 | Bacillales | H. glycines | SRR6269845 | | 0.1 | 13146 | 40 | C | 1236 | Gammaproteobacteria | H. glycines | SRR6269845 | | 0.09 | 10992 | 10992 | S | 2306583 | Halomonas sp. JS92-SW72 | H. glycines | SRR6269845 | | 0.07 | 8400 | 14 | G1 | 86661 | Bacillus cereus group | H. glycines | SRR6269845 | | 0.07 | 8529 | 8529 | S | 1034377 | Soybean cyst nematode socyvirus | H. glycines | SRR6269845 | | 0.07 | 8537 | 22 | G | 1386 | Bacillus | H. glycines | SRR6269845 | | 0.06 | 8344 | 8344 | S1 | 526973 | Bacillus cereus m1293 | H. glycines | SRR6269845 | | 0.06 | 8375 | 27 | S | 1396 | Bacillus cereus | H. glycines | SRR6269845 | | 0.04 | 5426 | 5426 | S | 1286 | Staphylococcus simulans | H. glycines | SRR6269845 | | 0.04 | 5601 | 128 | G | 1279 | Staphylococcus | H. glycines | SRR6269845 | | 0.02 | 2203 | 656 | G | 149698 | Massilia | H. glycines | SRR6269845 | | 0.02 | 2341 | 2341 | S | 2057025 | Spirosoma pollinicola | H. glycines | SRR6269845 | | 0.02 | 2742 | 65 | G | 107 | Spirosoma | H. glycines | SRR6269845 | | 0.02 | 2787 | 2787 | S1 | 395495 | Leptothrix cholodnii SP-6 | H. glycines | SRR6269845 | | 0.02 | 2825 | 231 | C | 28211 | Alphaproteobacteria | H. glycines | SRR6269845 | | 0.02 | 2836 | 271 | F | 75682 | Oxalobacteraceae | H. glycines | SRR6269845 | | 0.01 | 1742 | 1742 | S | 76731 | Roseateles depolymerans | H. glycines | SRR6269845 | | 0.01 | 1905 | 1905 | S | 1768242 | Paucibacter sp. KCTC 42545 | H. glycines | SRR6269845 | | 0.01 | 685 | 23 | F | 563835 | Chitinophagaceae | H. glycines | SRR6269845 | | 0.01 | 711 | 14 | O | 135614 | Xanthomonadales | H. glycines | SRR6269845 | | 0.01 | 764 | 764 | S | 1141883 | Massilia putida | H. glycines | SRR6269845 | | 0.01 | 776 | 308 | G | 12916 | Acidovorax | H. glycines | SRR6269845 | | 0.01 | 821 | 821 | S | 864051 | Burkholderiales bacterium JOSHI_001 | H. glycines | SRR6269845 | | 0.01 | 845 | 845 | S1 | 983917 | Rubrivivax gelatinosus IL144 | H. glycines | SRR6269845 | | 0.01 | 865 | 88 | O | 356 | Rhizobiales | H. glycines | SRR6269845 | | 0.01 | 869 | 23 | C | 1760 | Actinobacteria | H. glycines | SRR6269845 | | 0.01 | 933 | 128 | G | 283 | Comamonas | H. glycines | SRR6269845 |
Globodera pallida SRR7775195 "white potato cyst nematode"
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|---------|----------------------------------|----------------|------------| | 95.93 | 5053142 | 5435 | O3 | 33284 | Tylenchoidea | G. pallida | SRR7775195 | | 95.93 | 5053190 | 28 | O2 | 33283 | Tylenchomorpha | G. pallida | SRR7775195 | | 95.81 | 5047167 | 24809 | F1 | 33286 | Heteroderinae | G. pallida | SRR7775195 | | 1.82 | 95964 | 4043 | D | 2 | Bacteria | G. pallida | SRR7775195 | | 1.6 | 84529 | 9226 | P | 1224 | Proteobacteria | G. pallida | SRR7775195 | | 1.28 | 67199 | 7378 | C | 28216 | Betaproteobacteria | G. pallida | SRR7775195 | | 1.13 | 59594 | 18847 | O | 80840 | Burkholderiales | G. pallida | SRR7775195 | | 0.52 | 27242 | 11208 | G | 149698 | Massilia | G. pallida | SRR7775195 | | 0.52 | 27637 | 377 | F | 75682 | Oxalobacteraceae | G. pallida | SRR7775195 | | 0.29 | 15476 | 15476 | S | 1141883 | Massilia putida | G. pallida | SRR7775195 | | 0.23 | 11998 | 9765 | F | 119060 | Burkholderiaceae | G. pallida | SRR7775195 | | 0.14 | 7198 | 32 | D1 | 1783272 | Terrabacteria group | G. pallida | SRR7775195 | | 0.13 | 7033 | 119 | P | 1239 | Firmicutes | G. pallida | SRR7775195 | | 0.12 | 6531 | 6531 | U | 0 | unclassified | G. pallida | SRR7775195 | | 0.1 | 5066 | 234 | C | 1236 | Gammaproteobacteria | G. pallida | SRR7775195 | | 0.1 | 5300 | 5300 | S1 | 1239783 | Bacillus atrophaeus UCMB-5137 | G. pallida | SRR7775195 | | 0.05 | 2746 | 2746 | S | 2306583 | Halomonas sp. JS92-SW72 | G. pallida | SRR7775195 | | 0.05 | 2892 | 48 | C | 28211 | Alphaproteobacteria | G. pallida | SRR7775195 | | 0.04 | 1927 | 1927 | S | 1553431 | Mycoavidus cysteinexigens | G. pallida | SRR7775195 | | 0.03 | 1497 | 1443 | F | 135621 | Pseudomonadaceae | G. pallida | SRR7775195 | | 0.03 | 1538 | 1538 | S | 189426 | Paenibacillus odorifer | G. pallida | SRR7775195 | | 0.03 | 1753 | 365 | F | 41297 | Sphingomonadaceae | G. pallida | SRR7775195 | | 0.02 | 1009 | 1008 | G | 165697 | Sphingopyxis | G. pallida | SRR7775195 | | 0.02 | 819 | 271 | F | 80864 | Comamonadaceae | G. pallida | SRR7775195 | | 0.01 | 270 | 267 | F | 942 | Anaplasmataceae | G. pallida | SRR7775195 | | 0.01 | 375 | 375 | G | 13687 | Sphingomonas | G. pallida | SRR7775195 | | 0.01 | 375 | 375 | G | 41275 | Brevundimonas | G. pallida | SRR7775195 | | 0.01 | 498 | 498 | S1 | 365046 | Ramlibacter tataouinensis TTB310 | G. pallida | SRR7775195 | | 0.01 | 527 | 527 | S | 1707785 | Massilia sp. WG5 | G. pallida | SRR7775195 |
Ditylenchus destructor SRR79443144 "potato tuber nematode"
| Proportion of reads | Reads rooted to taxon clade | taxon-specific reads | rank code | Taxid | Taxon | Source Species | SRA | |---------------------|-----------------------------|----------------------|-----------|---------|---------------------------------|----------------|------------| | 74.57 | 38244277 | 20662 | O2 | 33283 | Tylenchomorpha | D.destructor | SRR7943144 | | 24.67 | 12650916 | 12650916 | U | 0 | unclassified | D.destructor | SRR7943144 | | 0.74 | 377285 | 24363 | D | 2 | Bacteria | D.destructor | SRR7943144 | | 0.43 | 222510 | 1353 | D1 | 1783272 | Terrabacteria group | D.destructor | SRR7943144 | | 0.38 | 195024 | 169 | P | 1239 | Firmicutes | D.destructor | SRR7943144 | | 0.36 | 186199 | 118 | C | 91061 | Bacilli | D.destructor | SRR7943144 | | 0.35 | 178437 | 200 | O | 1385 | Bacillales | D.destructor | SRR7943144 | | 0.34 | 173238 | 124 | G | 1386 | Bacillus | D.destructor | SRR7943144 | | 0.34 | 174009 | 11 | F | 186817 | Bacillaceae | D.destructor | SRR7943144 | | 0.33 | 170904 | 170904 | S1 | 526973 | Bacillus cereus m1293 | D.destructor | SRR7943144 | | 0.33 | 171059 | 126 | S | 1396 | Bacillus cereus | D.destructor | SRR7943144 | | 0.33 | 171332 | 106 | G1 | 86661 | Bacillus cereus group | D.destructor | SRR7943144 | | 0.26 | 131538 | 13212 | O3 | 33284 | Tylenchoidea | D.destructor | SRR7943144 | | 0.16 | 79642 | 3521 | P | 1224 | Proteobacteria | D.destructor | SRR7943144 | | 0.11 | 54016 | 14890 | F1 | 33286 | Heteroderinae | D.destructor | SRR7943144 | | 0.09 | 44143 | 19 | D2 | 68336 | Bacteroidetes/Chlorobi group | D.destructor | SRR7943144 | | 0.08 | 42879 | 562 | P | 976 | Bacteroidetes | D.destructor | SRR7943144 | | 0.07 | 36733 | 2516 | C | 1236 | Gammaproteobacteria | D.destructor | SRR7943144 | | 0.05 | 24443 | 24443 | S | 2057025 | Spirosoma pollinicola | D.destructor | SRR7943144 | | 0.05 | 25159 | 382 | F | 89373 | Cytophagaceae | D.destructor | SRR7943144 | | 0.04 | 21330 | 1063 | C | 28211 | Alphaproteobacteria | D.destructor | SRR7943144 | | 0.03 | 14857 | 480 | C | 1760 | Actinobacteria | D.destructor | SRR7943144 | | 0.02 | 10869 | 64 | O | 356 | Rhizobiales | D.destructor | SRR7943144 | | 0.02 | 8008 | 226 | P | 1117 | Cyanobacteria | D.destructor | SRR7943144 | | 0.02 | 8378 | 298 | F | 49546 | Flavobacteriaceae | D.destructor | SRR7943144 | | 0.02 | 8457 | 16 | O | 200644 | Flavobacteriales | D.destructor | SRR7943144 | | 0.02 | 9563 | 323 | C | 28216 | Betaproteobacteria | D.destructor | SRR7943144 | | 0.01 | 2627 | 2627 | S1 | 224013 | Nostoc piscinale CENA21 | D.destructor | SRR7943144 | | 0.01 | 2749 | 2749 | S | 95300 | Pseudomonas vancouverensis | D.destructor | SRR7943144 | | 0.01 | 2820 | 2820 | S | 2004644 | Acinetobacter sp. WCHA45 | D.destructor | SRR7943144 | | 0.01 | 2847 | 2847 | S | 1813019 | Campylobacter hepaticus | D.destructor | SRR7943144 | | 0.01 | 2866 | 29 | G | 1177 | Nostoc | D.destructor | SRR7943144 | | 0.01 | 2937 | 845 | G | 1883 | Streptomyces | D.destructor | SRR7943144 | | 0.01 | 3020 | 3020 | S | 2014542 | Alcanivorax sp. N3-2A | D.destructor | SRR7943144 | | 0.01 | 3057 | 17 | G | 59753 | Alcanivorax | D.destructor | SRR7943144 | | 0.01 | 3103 | 1014 | G | 662 | Vibrio | D.destructor | SRR7943144 | | 0.01 | 3415 | 224 | F | 641 | Vibrionaceae | D.destructor | SRR7943144 | | 0.01 | 3667 | 14 | F | 171552 | Prevotellaceae | D.destructor | SRR7943144 | | 0.01 | 3716 | 57 | O | 1161 | Nostocales | D.destructor | SRR7943144 | | 0.01 | 3804 | 3792 | S | 1491 | Clostridium botulinum | D.destructor | SRR7943144 | | 0.01 | 4261 | 17 | O | 171549 | Bacteroidales | D.destructor | SRR7943144 | | 0.01 | 4373 | 84 | G | 194 | Campylobacter | D.destructor | SRR7943144 | | 0.01 | 4496 | 142 | G | 469 | Acinetobacter | D.destructor | SRR7943144 | | 0.01 | 4761 | 29 | G | 1485 | Clostridium | D.destructor | SRR7943144 | | 0.01 | 4800 | 14 | F | 31979 | Clostridiaceae | D.destructor | SRR7943144 | | 0.01 | 4813 | 909 | F | 119060 | Burkholderiaceae | D.destructor | SRR7943144 | | 0.01 | 4880 | 18 | F | 2062 | Streptomycetaceae | D.destructor | SRR7943144 | | 0.01 | 5080 | 5080 | S1 | 316058 | Rhodopseudomonas palustris HaA2 | D.destructor | SRR7943144 | | 0.01 | 5879 | 746 | G | 286 | Pseudomonas | D.destructor | SRR7943144 | | 0.01 | 6121 | 36 | F | 135621 | Pseudomonadaceae | D.destructor | SRR7943144 | | 0.01 | 6586 | 59 | O | 80840 | Burkholderiales | D.destructor | SRR7943144 | | 0.01 | 6918 | 571 | O | 91347 | Enterobacterales | D.destructor | SRR7943144 | | 0.01 | 6952 | 22 | O | 186802 | Clostridiales | D.destructor | SRR7943144 | | 0.01 | 7644 | 149 | O | 186826 | Lactobacillales | D.destructor | SRR7943144 |
Network visualization in cytoscape
Here I summarized the summaries above by creating text files of two columns.
1
for f in *summary; do cut -f 6,7 $f >>${f%.*}.network;done
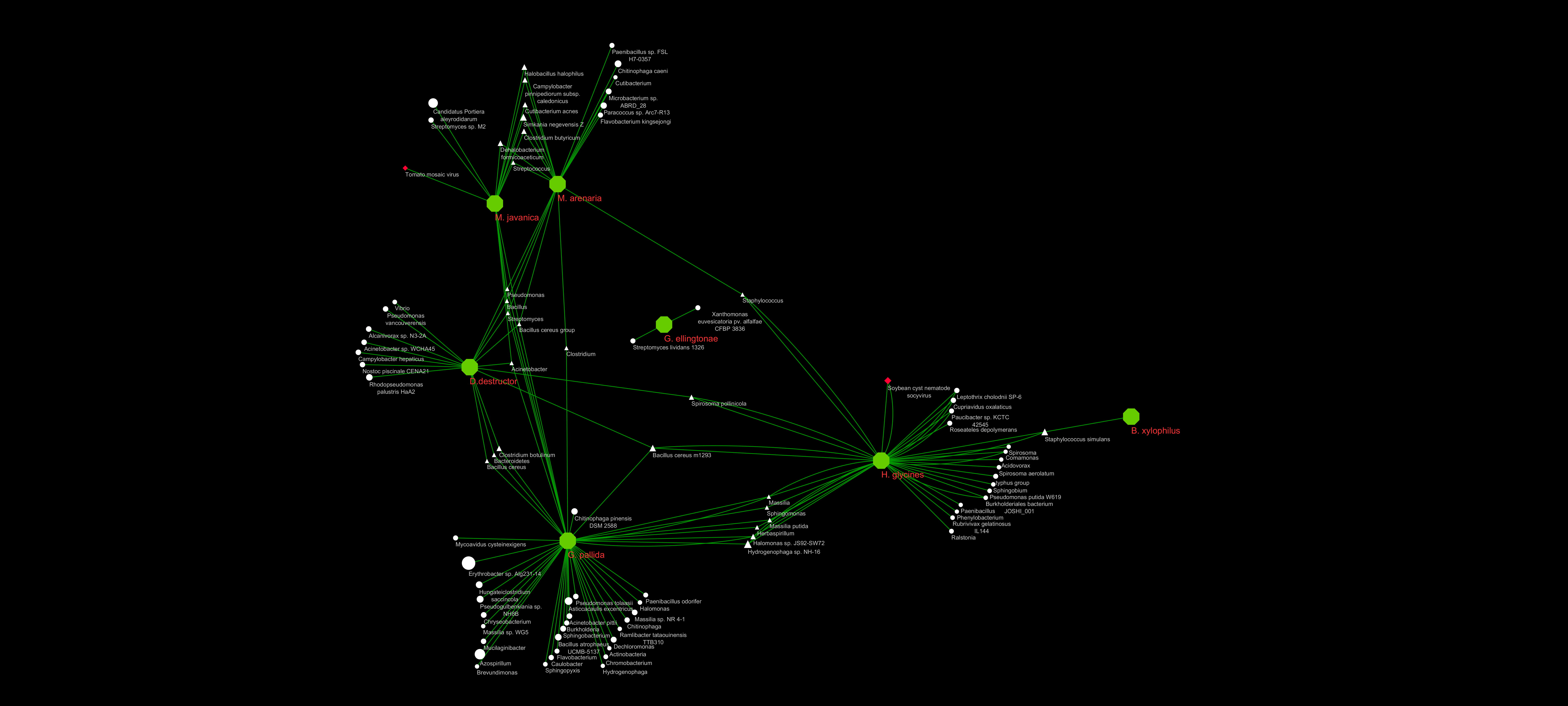
I labeled large green hexagons as the species, red diamonds as viruses, and triangles as bacteria present in two or more species.
Sources
1
2
3
The best source to get more information on kraken2
https://github.com/DerrickWood/kraken2/blob/master/docs/MANUAL.markdown
