Pyscaf is a genome scaffolding program that can scaffold assemblies based on synteny, long reads, and short reads.
Dependencies: Python 2.7,python FastaIndex, py-pillow, LAST assembler
Load the dependencies and needed files
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
| #Things needed to run pyscaf
git clone https://github.com/lpryszcz/pyScaf.git
module load python
module load py-fastaindex/0.11rc7-py2-sj3lhkk
module load last
module load py-pillow/3.2.0-py2-ods24od
#Need to be able to see what was done to each assembly also, so we need this.
This perl script will give us quick access to pertinent assembly stats.
git clone https://github.com/ucdavis-bioinformatics/assemblathon2-analysis.git
#get the two genomes you want to scaffold.
## I am just using a couple of E. coli genomes, because they will be small and quick to demonstrate the process.
wget ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/249/815/GCF_000249815.1_ASM24981v2/GCF_000249815.1_ASM24981v2_genomic.fna.gz
wget ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/732/965/GCF_000732965.1_ASM73296v1/GCF_000732965.1_ASM73296v1_genomic.fna.gz
gunzip GCF_000249815.1_ASM24981v2_genomic.fna.gz
gunzip GCF_000732965.1_ASM73296v1_genomic.fna.gz
#pyscaf will throw errors with a fna extension, so change those to .fa
mv GCF_000249815.1_ASM24981v2_genomic.fna GCF_000249815.1_ASM24981v2_genomic.fa
mv GCF_000732965.1_ASM73296v1_genomic.fna GCF_000732965.1_ASM73296v1_genomic.fa
|
Run pyscaf on the two E. coli genomes
1
2
3
4
5
6
7
8
| module load python
module load py-fastaindex/0.11rc7-py2-sj3lhkk
module load last
module load py-pillow/3.2.0-py2-ods24od
python pyScaf.py -f ../GCF_000249815.1_ASM24981v2_genomic.fa -t 16 --log ../E.coliPyscaf.output -r ../GCF_000732965.1_ASM73296v1_genomic.fa
#this analysis took 5 seconds!
|
What are the changes in the genome stats?
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
| cd /work/GIF/remkv6/USDA/16_pyscaf/assemblathon2-analysis
perl assemblathon_stats.pl ../GCF_000249815.1_ASM24981v2_genomic.fa
##############################################################################
---------------- Information for assembly '../GCF_000249815.1_ASM24981v2_genomic.fa' ----------------
Number of scaffolds 100
Total size of scaffolds 5371046
Longest scaffold 306092
Shortest scaffold 301
Number of scaffolds > 1K nt 90 90.0%
Number of scaffolds > 10K nt 62 62.0%
Number of scaffolds > 100K nt 22 22.0%
Number of scaffolds > 1M nt 0 0.0%
Number of scaffolds > 10M nt 0 0.0%
Mean scaffold size 53710
Median scaffold size 18594
N50 scaffold length 133903
L50 scaffold count 15
scaffold %A 24.72
scaffold %C 25.29
scaffold %G 25.33
scaffold %T 24.65
scaffold %N 0.00
scaffold %non-ACGTN 0.00
Number of scaffold non-ACGTN nt 0
Percentage of assembly in scaffolded contigs 0.0%
Percentage of assembly in unscaffolded contigs 100.0%
Average number of contigs per scaffold 1.0
Average length of break (>25 Ns) between contigs in scaffold 0
Number of contigs 100
Number of contigs in scaffolds 0
Number of contigs not in scaffolds 100
Total size of contigs 5371046
Longest contig 306092
Shortest contig 301
Number of contigs > 1K nt 90 90.0%
Number of contigs > 10K nt 62 62.0%
Number of contigs > 100K nt 22 22.0%
Number of contigs > 1M nt 0 0.0%
Number of contigs > 10M nt 0 0.0%
Mean contig size 53710
Median contig size 18594
N50 contig length 133903
L50 contig count 15
contig %A 24.72
contig %C 25.29
contig %G 25.33
contig %T 24.65
contig %N 0.00
contig %non-ACGTN 0.00
Number of contig non-ACGTN nt 0
###############################################################################
perl assemblathon_stats.pl ../scaffolds.fa
###############################################################################
---------------- Information for assembly '../scaffolds.fa' ----------------
Number of scaffolds 61
Total size of scaffolds 5545685
Longest scaffold 707086
Shortest scaffold 301
Number of scaffolds > 1K nt 57 93.4%
Number of scaffolds > 10K nt 35 57.4%
Number of scaffolds > 100K nt 17 27.9%
Number of scaffolds > 1M nt 0 0.0%
Number of scaffolds > 10M nt 0 0.0%
Mean scaffold size 90913
Median scaffold size 16148
N50 scaffold length 256789
L50 scaffold count 6
scaffold %A 23.81
scaffold %C 24.43
scaffold %G 24.38
scaffold %T 23.78
scaffold %N 3.61
scaffold %non-ACGTN 0.00
Number of scaffold non-ACGTN nt 0
Percentage of assembly in scaffolded contigs 63.5%
Percentage of assembly in unscaffolded contigs 36.5%
Average number of contigs per scaffold 1.5
Average length of break (>25 Ns) between contigs in scaffold 6450
Number of contigs 92
Number of contigs in scaffolds 40
Number of contigs not in scaffolds 52
Total size of contigs 5345715
Longest contig 428031
Shortest contig 301
Number of contigs > 1K nt 82 89.1%
Number of contigs > 10K nt 55 59.8%
Number of contigs > 100K nt 22 23.9%
Number of contigs > 1M nt 0 0.0%
Number of contigs > 10M nt 0 0.0%
Mean contig size 58106
Median contig size 17067
N50 contig length 144532
L50 contig count 12
contig %A 24.70
contig %C 25.34
contig %G 25.29
contig %T 24.67
contig %N 0.00
contig %non-ACGTN 0.00
Number of contig non-ACGTN nt 0
##############################################################################
Pyscaf also provides a standard output log of what was scaffolded in a tsv file (scaffolds.fa.tsv). Here is just what changed, not the entire output.
##############################################################################
# name size no. of contigs ordered contigs contig orientations (0-forward; 1-reverse) gap sizes (negative gap size = adjacent contigs are overlapping)
scaffold00001 256789 3 NZ_AIGG01000001.1 NZ_AIGG01000095.1 NZ_AIGG01000002.1 0 1 0 4665 51 0
scaffold00002 521794 4 NZ_AIGG01000004.1 NZ_AIGG01000005.1 NZ_AIGG01000006.1 NZ_AIGG01000008.1 0 0 0 0 8114 1201 2221 0
scaffold00003 16148 2 NZ_AIGG01000014.1 NZ_AIGG01000015.1 0 0 -3019 0
scaffold00004 562870 4 NZ_AIGG01000018.1 NZ_AIGG01000025.1 NZ_AIGG01000027.1 NZ_AIGG01000010.1 0 0 1 1 8969 -14857 8327 0
scaffold00005 69957 4 NZ_AIGG01000092.1 NZ_AIGG01000094.1 NZ_AIGG01000100.1 NZ_AIGG01000039.1 0 0 0 0 150 502 14358 0
scaffold00006 707086 5 NZ_AIGG01000045.1 NZ_AIGG01000050.1 NZ_AIGG01000051.1 NZ_AIGG01000053.1 NZ_AIGG01000054.1 0 0 0 0 0 29777 -15 37697 5 0
scaffold00007 227254 4 NZ_AIGG01000056.1 NZ_AIGG01000057.1 NZ_AIGG01000058.1 NZ_AIGG01000059.1 0 0 0 0 162 264 386 0
scaffold00008 76958 2 NZ_AIGG01000067.1 NZ_AIGG01000068.1 0 0 -5134 0
scaffold00009 577718 11 NZ_AIGG01000035.1 NZ_AIGG01000070.1 NZ_AIGG01000072.1 NZ_AIGG01000073.1 NZ_AIGG01000075.1 NZ_AIGG01000076.1 NZ_AIGG01000077.1 NZ_AIGG01000062.1 NZ_AIGG01000063.1 NZ_AIGG01000064.1 NZ_AIGG01000065.1 1 0 0 0 0 0 0 0 0 0 0 19075 1819 660 2012 146 182 43604 273 73 182 0
scaffold00010 355331 7 NZ_AIGG01000079.1 NZ_AIGG01000080.1 NZ_AIGG01000096.1 NZ_AIGG01000081.1 NZ_AIGG01000074.1 NZ_AIGG01000082.1 NZ_AIGG01000069.1 0 0 0 0 0 0 1 322 28 110 -348 -2 5278 0
scaffold00011 242324 4 NZ_AIGG01000086.1 NZ_AIGG01000087.1 NZ_AIGG01000089.1 NZ_AIGG01000090.1 0 0 1 0 28 9334 -1961 0
##############################################################################
|
There is also a nice dotplot output if there aren’t too many scaffolds.
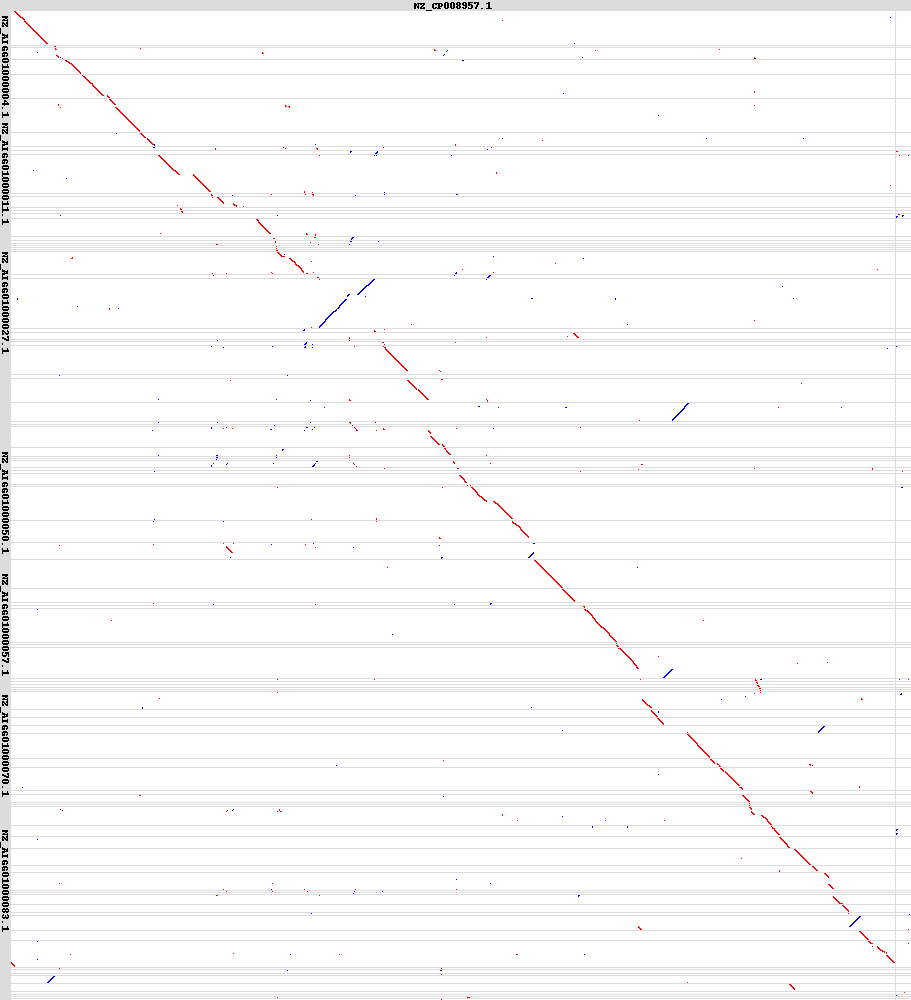
PyScaf also works on larger genomes rather quickly
Though genomes at the size of 1Gbp the scaffolding decreases in speed significantly. Here I’ll scaffold a fragmented Arabidopsis genome assembly with a chromosome level Arabidopsis genome assembly.
Get genomes
1
2
3
4
5
6
7
8
9
10
11
12
13
14
| #here is chromosome level assembly of arabidosis thaliana
wget ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/001/735/GCF_000001735.4_TAIR10.1/GCF_000001735.4_TAIR10.1_genomic.fna.gz
Here is an arabidopsis thaliana assembly that is in the thousands of scaffolds
wget ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/001/742/845/GCA_001742845.1_AthNd1_v1.0/GCA_001742845.1_AthNd1_v1.0_genomic.fna.gz
gunzip GCF_000001735.4_TAIR10.1_genomic.fna.gz
gunzip GCA_001742845.1_AthNd1_v1.0_genomic.fna.gz
# fix the file extensions so they are fa, not fna
mv GCA_001742845.1_AthNd1_v1.0_genomic.fna GCA_001742845.1_AthNd1_v1.0_genomic.fa
mv GCF_000001735.4_TAIR10.1_genomic.fna GCF_000001735.4_TAIR10.1_genomic.fa
|
Run pyscaf on arabidopsis genomes
1
2
3
| #this took 21 minutes to finish using 16 processors.
python pyScaf.py -f ../01_Arabidopsis/GCA_001742845.1_AthNd1_v1.0_genomic.fa -t 16 --log ../01_Arabidopsis/ArabidopsisPyscaf.output -r ../01_Arabidopsis/GCF_000001735.4_TAIR10.1_genomic
.fa
|
Output of pyscaf
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
| #Assembly prior to scaffolding
perl assemblathon_stats.pl ../01_Arabidopsis/GCA_001742845.1_AthNd1_v1.0_genomic.fa
##############################################################################
---------------- Information for assembly '../01_Arabidopsis/GCA_001742845.1_AthNd1_v1.0_genomic.fa' ----------------
Number of scaffolds 5197
Total size of scaffolds 116846015
Longest scaffold 2967516
Shortest scaffold 500
Number of scaffolds > 1K nt 2220 42.7%
Number of scaffolds > 10K nt 630 12.1%
Number of scaffolds > 100K nt 205 3.9%
Number of scaffolds > 1M nt 26 0.5%
Number of scaffolds > 10M nt 0 0.0%
Mean scaffold size 22483
Median scaffold size 836
N50 scaffold length 589639
L50 scaffold count 49
scaffold %A 31.71
scaffold %C 17.79
scaffold %G 17.78
scaffold %T 31.67
scaffold %N 1.04
scaffold %non-ACGTN 0.00
Number of scaffold non-ACGTN nt 0
Percentage of assembly in scaffolded contigs 77.2%
Percentage of assembly in unscaffolded contigs 22.8%
Average number of contigs per scaffold 1.2
Average length of break (>25 Ns) between contigs in scaffold 1468
Number of contigs 6021
Number of contigs in scaffolds 1232
Number of contigs not in scaffolds 4789
Total size of contigs 115635978
Longest contig 2204473
Shortest contig 285
Number of contigs > 1K nt 2987 49.6%
Number of contigs > 10K nt 853 14.2%
Number of contigs > 100K nt 278 4.6%
Number of contigs > 1M nt 8 0.1%
Number of contigs > 10M nt 0 0.0%
Mean contig size 19205
Median contig size 985
N50 contig length 263174
L50 contig count 110
contig %A 32.04
contig %C 17.98
contig %G 17.97
contig %T 32.00
contig %N 0.01
contig %non-ACGTN 0.00
Number of contig non-ACGTN nt 0
##############################################################################
perl assemblathon_stats.pl ../pyScaf/scaffolds.fa
##############################################################################
---------------- Information for assembly '../pyScaf/scaffolds.fa' ----------------
Number of scaffolds 4863
Total size of scaffolds 119654142
Longest scaffold 22790922
Shortest scaffold 500
Number of scaffolds > 1K nt 1974 40.6%
Number of scaffolds > 10K nt 481 9.9%
Number of scaffolds > 100K nt 100 2.1%
Number of scaffolds > 1M nt 6 0.1%
Number of scaffolds > 10M nt 5 0.1%
Mean scaffold size 24605
Median scaffold size 804
N50 scaffold length 12441124
L50 scaffold count 4
scaffold %A 29.87
scaffold %C 16.77
scaffold %G 16.75
scaffold %T 29.84
scaffold %N 6.77
scaffold %non-ACGTN 0.00
Number of scaffold non-ACGTN nt 0
Percentage of assembly in scaffolded contigs 88.3%
Percentage of assembly in unscaffolded contigs 11.7%
Average number of contigs per scaffold 1.2
Average length of break (>25 Ns) between contigs in scaffold 7983
Number of contigs 5876
Number of contigs in scaffolds 1329
Number of contigs not in scaffolds 4547
Total size of contigs 111566809
Longest contig 2394530
Shortest contig 285
Number of contigs > 1K nt 2884 49.1%
Number of contigs > 10K nt 799 13.6%
Number of contigs > 100K nt 257 4.4%
Number of contigs > 1M nt 10 0.2%
Number of contigs > 10M nt 0 0.0%
Mean contig size 18987
Median contig size 967
N50 contig length 284383
L50 contig count 96
contig %A 32.04
contig %C 17.98
contig %G 17.97
contig %T 32.00
contig %N 0.01
contig %non-ACGTN 0.00
Number of contig non-ACGTN nt 0
##############################################################################
This time pyscaf was able to scaffold near ~350 scaffolds. This may not sound incredible, but when using a chromosome level assembly for scaffolding, this can be tremendousl
##############################################################################
# name size no. of contigs ordered contigs contig orientations (0-forward; 1-reverse) gap sizes (negative gap size = adjacent contigs are overlapping)
scaffold00001 22790922 108 KV766200.1 KV766201.1 LXSY01000010.1 LXSY01000011.1 LXSY01000012.1 LXSY01000013.1 KV766202.1 LXSY01000021.1 LXSY01000022.1 LXSY01000023.1 LXSY01000027.1 LXSY01000028.1 KV766203.1 LXSY01000037.1 LXSY01000038.1 LXSY01000039.1 LXSY01000040.1 LXSY01000041.1 KV766205.1 KV766206.1 LXSY01000052.1 LXSY01000053.1 KV766207.1 LXSY01000066.1 LXSY01000067.1 LXSY01000068.1 KV766208.1 KV766209.1 LXSY01000080.1 LXSY01000081.1 LXSY01000082.1 LXSY01000083.1 KV766210.1 LXSY01000089.1 KV766211.1 KV766212.1 KV766213.1 LXSY01000103.1 LXSY01000104.1 LXSY01000105.1 LXSY01000106.1 KV766214.1 KV766215.1 KV766217.1 LXSY01000134.1 LXSY01000135.1 KV766218.1 LXSY01000141.1 LXSY01000142.1 LXSY01000143.1 LXSY01000144.1 LXSY01000145.1 LXSY01000146.1 KV766219.1 LXSY01000154.1 KV766220.1 LXSY01000165.1 LXSY01000166.1 LXSY01000167.1 LXSY01000168.1 LXSY01000170.1 LXSY01000171.1 LXSY01000172.1 KV766240.1 KV766256.1 LXSY01001087.1 KV766281.1 LXSY01001187.1 KV766282.1 KV766286.1 LXSY01001256.1 LXSY01001257.1 LXSY01001258.1 LXSY01001259.1 LXSY01001270.1 LXSY01001272.1 LXSY01001273.1 KV766289.1 KV766290.1 KV766291.1 LXSY01001312.1 LXSY01001313.1 KV766292.1 LXSY01001318.1 LXSY01001319.1 LXSY01001326.1 KV766295.1 KV766296.1 LXSY01001356.1 KV766298.1 LXSY01001372.1 LXSY01001374.1 KV766299.1 LXSY01001384.1 LXSY01001385.1 LXSY01001386.1 KV766300.1 KV766301.1 LXSY01001418.1 LXSY01001419.1 LXSY01001420.1 LXSY01001421.1 LXSY01001422.1 KV766302.1 LXSY01001435.1 LXSY01001436.1 LXSY01001437.1 KV766303.1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 -5 116 240 4 3 11337 3 208 -3569 -1356 4 -18 -73 -15 -12 283 -12 2213 1 509 315 716 -533 4 5 -11 -12 -104 66 4980 211 -39 -16 1879 -4 20 123 51 -2 1753 -4 3894 -30446 20493 694 98 -5 -9 16 63 -27 -372 49 468 379 2100 -425 436 -703 2733 5 879 734063 -1699 24576 298 644 7423 -5356 -49450 -5 11048 109 6 5 47 -221 -124168 -519806 -23 -8 48 632 6 39965 201635 21 1130 -380 -4688 -1899 571 2124 -16 -104 -337916 -7495 2502 -17 30 2455 803 375 -3 637 340 1189 0
scaffold00002 16660361 81 KV766583.1 LXSY01004999.1 LXSY01005000.1 KV766584.1 LXSY01005006.1 LXSY01005007.1 KV766585.1 LXSY01005019.1 KV766586.1 KV766587.1 KV766589.1 KV766592.1 KV766593.1 LXSY01005104.1 LXSY01005110.1 LXSY01005111.1 LXSY01005113.1 LXSY01005115.1 LXSY01005401.1 LXSY01005402.1 LXSY01005404.1 LXSY01005407.1 LXSY01005408.1 LXSY01005412.1 LXSY01005414.1 LXSY01005415.1 LXSY01005419.1 LXSY01005423.1 KV766625.1 LXSY01005431.1 LXSY01005432.1 LXSY01005437.1 LXSY01005462.1 LXSY01005441.1 KV766624.1 LXSY01005485.1 LXSY01005491.1 LXSY01005492.1 LXSY01005494.1 LXSY01003304.1 LXSY01004332.1 LXSY01005516.1 LXSY01005518.1 LXSY01005538.1 LXSY01003103.1 LXSY01005457.1 LXSY01005546.1 LXSY01005547.1 LXSY01005549.1 KV766631.1 LXSY01005600.1 LXSY01005602.1 LXSY01005612.1 LXSY01005613.1 KV766633.1 LXSY01005614.1 LXSY01005615.1 LXSY01005616.1 LXSY01005617.1 LXSY01005620.1 LXSY01005623.1 LXSY01005626.1 LXSY01005627.1 LXSY01005660.1 KV766636.1 LXSY01005761.1 KV766672.1 LXSY01006146.1 KV766686.1 KV766687.1 LXSY01006159.1 LXSY01006160.1 KV766688.1 LXSY01006178.1 LXSY01006179.1 LXSY01006180.1 KV766689.1 KV766691.1 KV766693.1 KV766694.1 KV766695.1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 656 18 1926 420 217 37 73 296 -906204 -49418 75872 169 65282 16130 252 34425 4912 404287 956 3669 19 1 7247 1470 89 15425 175 -413 -456 -467 -362 -331 1393 272 -93 -257 -380 1417 -727 -836 -182 1937 761 -103 -318 1970 3077 8389 913 6936 6371 38416 2 1132 1152 -360 425 -1031 142 566 1228 -17 -231 2058 405485 -277297 904536 -6 78 74 78 310 -17 3077 -17 260 -469413 -409 1559 700 0
scaffold00003 15040960 43 KV766389.1 KV766390.1 KV766391.1 KV766392.1 LXSY01002537.1 LXSY01002538.1 LXSY01002539.1 KV766393.1 KV766394.1 KV766395.1 KV766396.1 KV766397.1 LXSY01002587.1 LXSY01002588.1 KV766398.1 LXSY01002597.1 KV766399.1 KV766400.1 LXSY01002623.1 KV766462.1 LXSY01003591.1 LXSY01003592.1 KV766472.1 LXSY01003597.1 LXSY01003598.1 LXSY01003599.1 LXSY01003600.1 LXSY01003601.1 KV766475.1 LXSY01003643.1 KV766480.1 KV766483.1 KV766485.1 LXSY01003682.1 LXSY01003683.1 LXSY01003684.1 LXSY01003685.1 LXSY01003686.1 KV766486.1 KV766488.1 KV766489.1 KV766490.1 KV766491.1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 -2341 100 1595 600 191 135 69 -5541 -67354 26 -639 1412 5076 298 1819 12346 1662 5098 1118312 109538 37782 11 151713 95 59802 3544 55494 70480 88913 7 -106105 -519006 -11 1345 289 329 432 1462 40591 -10 285 0 0
scaffold00004 12441124 45 LXSY01001453.1 LXSY01001454.1 LXSY01001455.1 LXSY01001456.1 LXSY01001621.1 LXSY01001462.1 LXSY01001466.1 KV766304.1 LXSY01001471.1 LXSY01001472.1 LXSY01001473.1 LXSY01001474.1 KV766305.1 LXSY01001480.1 KV766306.1 KV766312.1 LXSY01002297.1 LXSY01002311.1 LXSY01002312.1 LXSY01002313.1 KV766374.1 LXSY01002317.1 KV766375.1 KV766378.1 KV766381.1 KV766382.1 LXSY01002408.1 LXSY01002409.1 LXSY01002410.1 LXSY01002411.1 LXSY01002412.1 KV766383.1 LXSY01002420.1 KV766384.1 LXSY01002425.1 KV766385.1 LXSY01002431.1 LXSY01002432.1 KV766386.1 KV766387.1 LXSY01002468.1 LXSY01002469.1 LXSY01002470.1 LXSY01002471.1 KV766388.1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1983 1935 -17 942 12633 7088 2984 136 288 458 353 279 251 148 -9798 153219 467493 5131 -9 1339 -16 77 -318956 285834 -296801 -185 201 606 441 -15 -17 4066 1046 202 1274 170 170 318 545 185 1281 1367 309 309 0
scaffold00005 12054261 62 KV766492.1 KV766494.1 LXSY01004106.1 LXSY01004111.1 LXSY01004113.1 LXSY01004114.1 KV766638.1 LXSY01004116.1 LXSY01004117.1 LXSY01004118.1 LXSY01004121.1 LXSY01004129.1 KV766517.1 LXSY01004132.1 LXSY01004133.1 LXSY01004134.1 LXSY01004135.1 KV766518.1 KV766543.1 LXSY01004733.1 LXSY01004798.1 LXSY01004799.1 LXSY01004800.1 LXSY01004801.1 LXSY01004802.1 LXSY01004803.1 KV766559.1 KV766562.1 LXSY01004845.1 KV766563.1 LXSY01004854.1 KV766565.1 KV766566.1 KV766567.1 KV766568.1 KV766570.1 LXSY01004893.1 KV766571.1 KV766572.1 LXSY01004905.1 LXSY01004906.1 LXSY01004908.1 KV766573.1 KV766574.1 LXSY01004919.1 LXSY01004920.1 KV766575.1 LXSY01004925.1 KV766576.1 LXSY01004928.1 KV766577.1 LXSY01004933.1 LXSY01004934.1 LXSY01004935.1 LXSY01004937.1 LXSY01004939.1 KV766578.1 LXSY01004947.1 KV766579.1 LXSY01004964.1 KV766582.1 LXSY01004990.1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 546842 -2099 4955 233 -171 -402 -411 -588 171 340 23250 3763 715 -170 4185 1059 2304 -16066 776 202411 3243 537 237 3491 -1 204133 -61959 1017 -7 19467 161 40 3 665 -170610 366 1062 1741 -34 585 105 917 -32640 -335 5442 -3 -61 -35 -13 194 -980 2203 41 470 2969 4568 19 -8 372 666 862 0
###############################################################################
So 108 scaffolds put into chromosome 1, 81 into chromosome 2, 43 into chromosome 3, 45 into chromosome 4, 62 into chromosome 5.
#unfortunately too many scaffolds to show in the dotplot, but a figure can be drawn by removing some of the small un-scaffoldable scaffolds from the assembly prior to running pyscaf.
|
Further reading

