Opscan 1:1 Ortholog Calling to Synteny with iAdhore
Opscan is a software that will call orthologous genes between two sets of genes, a prerequisite for constructing synteny between species. There are many other software options out there that can provide ortholog calling capabilities, including BLAST, OrthoMCL, OrthoFinder, etc. However, Opscan offers some unique benefits and is extremely fast. (20mins on 30k genes) Opscan was published was also published as part of a synteny software, thus corroborating its use for such Opscan Publication.
Synopsis from Opscan manual
“Fastp then NWS (without end gap penalties) operon search in genome database starting with an operon file. Can be used with a genome database instead of an operon in genome versus genome comparisons.””
Download the program files
1
2
3
4
5
#available here http://www.lcqb.upmc.fr/CHROnicle/SynChro.html
wget http://www.lcqb.upmc.fr/CHROnicle/telechargements/SynChro_linux.zip
unzip SynChro_linux.unzip
The relevant program file can be found in the downloaded folder directory
/1SynChro/Opscan/bin/opscan
Create the input files
Opscan uses two files that you will need to create from a predicted protein fasta file of the genes and a gff annotating gene positions in the genome.
1
2
3
4
5
6
:Usage of opscan
Opscan [options] -O <operon.db> [<genome.db>]
:Files that need to be created from the gff and and protein fasta for each species
1. operon.db
2. genome.db
Acquire your gff and gene fasta.
Note: it is good to make sure that no repetitive elements (Transposons,etc) are not called in your gene set. This can cause downstream errors with synteny.
Sample 1 protein fasta
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
>GPLIN_000192700 transcript_id=GPLIN_000192700 gene_id=GPLIN_000192700
MSGRGRGGGFVPKKRLNISNQIGINESQQMTAIIQKDEQPKKDMQKDEQSKKDIQKDEQS
KKDIQKDELPKKEESKESLEKQPAERLQQQHPPPPIVHHNQKEHHHIVARKQPVVCFYCK
YEGHMLADCEKRMHMEAKKKAAQIVPRVVNYTFRRTTSQERSGGVGAVSAVTVSPNENSS
FDVFTKPYEGGQSEDDDVATGEDDEEKENDVNEEEKKNAEEERGKDGGKEEQKDEEGTGD
EHHRQQKQEQPLKDVVAAGLVNDGQQEQHQRKKATTASSKKPLVVVPLPVPVSTVKAQVQ
LRSKLLSSNSAGGSPAATSPVVVNPRSESSEEEDDEEVVETPEEDNGSCGEEVGGQSKTI
VTSSATASEETAPPKTEGTEGLEKLVQKLNMDEVDTQDEHLQPEKGGGEDELETVSVKVD
GCVEFDSDSSSYYDCSDGEGGGDGRRLKRIVEEDEYYGEHGALLDLEDDEPAVVGLAAEA
VDTKNEGEEPQQEEGAETDSY
>GPLIN_001029200 transcript_id=GPLIN_001029200 gene_id=GPLIN_001029200
MALVNGKCKHFCRLIKCWLHELAIMKMEQKQYIKNFNFVLKLSEFSLSYYEVYKVLEAIL
IESNAASAIITNERPPAVHTFLKQHFISKLYRKSIKLKQYIRLKFGSNMNKVDCAFVHFG
REMERRLEENSFISYPRFSEYVEQMEEEKCGEAPHNQNTQNTTDLGSKQLAMKLFHEPLW
NYARAIQIDSHECKAVLQFADALRP
>GPLIN_001497500 transcript_id=GPLIN_001497500 gene_id=GPLIN_001497500
MVENATVAATTTDFPPTPTGSTPEGPLNRMGNFLAGLVGGEHQQQLQQTTTPGANASDHE
ALSNNDRVANASSTVSANIDAGGIATTVTPPLDNTTSNVFSSTTEMSPSIVEGPEVEVWN
FAEGMDNMTTPSSTSSTTTERPPVPASAAVALNAWAVNLVASAAVVAVVFFRSF
>GPLIN_001590800 transcript_id=GPLIN_001590800 gene_id=GPLIN_001590800
MKGRWRPANSEHAVTEQVVVRDGGRRKKSDERHWGASARGPAERAHGDPQPGHARTSSGA
RSLAPPTGRRSRVCGTLAAIFHLTV
>GPLIN_000513900 transcript_id=GPLIN_000513900 gene_id=GPLIN_000513900
MEEVAASTPTSRDQSQTRHIRHRKGSDHFEPMYNQQLFVNIENSIGGSAHQSTSNLMHSP
KNVFENGQSQLVLELIGTEEEKPRYYSMPPEVQSEPAVIKLMQSGKKLHIYEGHIFVAVK
PRGRDNMDVYKQQHFQHEHRGSGLARILEAQPS
>GPLIN_000179500 transcript_id=GPLIN_000179500 gene_id=GPLIN_000179500
MSDNPKKVEKRLKEIFICDDVLFAVFKFCGPFELGLKVALISDRFDLLVDAHFKSMEWSL
GCLEIHSATDGSGAEIVKCLSNNGQVERRLPIPQEALPDNVIGFKRLEISYMDQSVIEFF
YRIRRLFASNGISLLISTKTNQKRSWEIIWQRIWPLINDNICGLFSLFPYNLERLRRFSP
TILRDCAKLRMIYSNNLFLKLSADDSAGASSEQALAKWLHTPRGDGLPKEFDNSAEPVNF
IIRFWHCYSDDIMPFELKNILTGERLVLRQINDFEWLLVRCRIERDEDKWTEWENKAVEG
DWRQRFAILFRDSAIGDG
>GPLIN_000089400 transcript_id=GPLIN_000089400 gene_id=GPLIN_000089400
MQQWGTSDIWKGLELDEHDCHAYFDGCQTYSCVDENGDQIFVVNSCVPDIGNCTHSDLDS
ICTKDGGTPKCEICYNGLCNKAKIELKKPLLKTVQKPNNPPETGTGGAKDKSSSEENGMT
TGAKNESSSEKNGMTTGAKNESSSEENGTTGAKASSTPAD
>GPLIN_000832600 transcript_id=GPLIN_000832600 gene_id=GPLIN_000832600
MLSNNFSILFVLLLVLIENNFIFAKSTRTPRTTTATASDEELDEIVNSISETPKNDGDDN
LSTSNEIFELGANSPTTNNGKNQYSASSHDSDEQQQQLALPKMRTEGNFTILPPPILTTE
QIDAIARQSAAMITRSALKHSKEIFAKFAPDSWTEALADVPLSTIRCTVRVSTKVRLLQQ
QAFLHEHLSFTLNNENEDDDTEADVISDGSDRETTTTTTTSNSNGGTPFGAEEASEIRSH
DQAAKTINETIRQLPPHVRQTLEEYGKRFKNQSLYLGGAEAVASIKSILVDLLEELKSVT
SEEREQLGDLFPKLRNVVTPLHTPV
>GPLIN_000567000 transcript_id=GPLIN_000567000 gene_id=GPLIN_000567000
MKTNKRQKVSSSEEEFDPNQKGEDYDLFMDNEEREKFDKMTEKEREIEIYKRLEQREIMK
TREKIKKKLEMAAGKKADEEGSSSQKKRKRKISEESNEGNDDEKEDSPSEAGEVDSSSDE
PKESSSSKGPVKAEPDESDGEIDADYHKPSEVANKQSKKKAMAELLSKRRNKRQTEEKRK
KETLKATLDLDEVFGNEDEEGKSASSSSSSTSSSRSSSPDLGKSRSPSPVAKQEIHTRDE
LIRCQLFRRKLAKFVHAPFFAKTVVGCFVRISIGPNKLDGKQVYRVVEVVETAKIYEVEP
NSDIRTNKGFRLKVCDGEPMRVYRLEFISNSHIVESEFLFWLNKLRSTNTPVPTVDFVDR
KRKEIVDALIYNYNESEVNLLIKEKQRFKRQPTNFAFQKAELLKIKVGG
etc.
Sample 1 genome gff
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
##gff-version 3
##sequence-region pathogens_Gpal_scaffold_1 1 599721
# Gene gene:GPLIN_000000100
pathogens_Gpal_scaffold_1 WormBase_imported gene 32 1060 . + . ID=gene:GPLIN_000000100;Name=GPLIN_000000100;biotype=protein_coding
pathogens_Gpal_scaffold_1 WormBase_imported mRNA 32 1060 . + . ID=transcript:GPLIN_000000100;Parent=gene:GPLIN_000000100;Name=GPLIN_000000100
pathogens_Gpal_scaffold_1 WormBase_imported exon 32 166 . + . ID=exon:GPLIN_000000100.1;Parent=transcript:GPLIN_000000100
pathogens_Gpal_scaffold_1 WormBase_imported exon 483 585 . + . ID=exon:GPLIN_000000100.2;Parent=transcript:GPLIN_000000100
pathogens_Gpal_scaffold_1 WormBase_imported exon 685 792 . + . ID=exon:GPLIN_000000100.3;Parent=transcript:GPLIN_000000100
pathogens_Gpal_scaffold_1 WormBase_imported exon 837 1060 . + . ID=exon:GPLIN_000000100.4;Parent=transcript:GPLIN_000000100
pathogens_Gpal_scaffold_1 WormBase_imported CDS 32 166 . + 0 ID=cds:GPLIN_000000100;Parent=transcript:GPLIN_000000100
pathogens_Gpal_scaffold_1 WormBase_imported CDS 483 585 . + 0 ID=cds:GPLIN_000000100;Parent=transcript:GPLIN_000000100
pathogens_Gpal_scaffold_1 WormBase_imported CDS 685 792 . + 2 ID=cds:GPLIN_000000100;Parent=transcript:GPLIN_000000100
pathogens_Gpal_scaffold_1 WormBase_imported CDS 837 1060 . + 2 ID=cds:GPLIN_000000100;Parent=transcript:GPLIN_000000100
###
# Gene gene:GPLIN_000000200
pathogens_Gpal_scaffold_1 WormBase_imported gene 47003 48028 . + . ID=gene:GPLIN_000000200;Name=GPLIN_000000200;biotype=protein_coding
pathogens_Gpal_scaffold_1 WormBase_imported mRNA 47003 48028 . + . ID=transcript:GPLIN_000000200;Parent=gene:GPLIN_000000200;Name=GPLIN_000000200;info=method:InterPro accession:IPR012677 description:Nucleotide-binding%2C alpha-beta plait %0Amethod:InterPro accession:IPR015047 description:Domain of unknown function DUF1866
pathogens_Gpal_scaffold_1 WormBase_imported exon 47003 47013 . + . ID=exon:GPLIN_000000200.1;Parent=transcript:GPLIN_000000200
pathogens_Gpal_scaffold_1 WormBase_imported exon 47446 47475 . + . ID=exon:GPLIN_000000200.2;Parent=transcript:GPLIN_000000200
pathogens_Gpal_scaffold_1 WormBase_imported exon 47527 47655 . + . ID=exon:GPLIN_000000200.3;Parent=transcript:GPLIN_000000200
pathogens_Gpal_scaffold_1 WormBase_imported exon 47711 47888 . + . ID=exon:GPLIN_000000200.4;Parent=transcript:GPLIN_000000200
pathogens_Gpal_scaffold_1 WormBase_imported exon 47942 48028 . + . ID=exon:GPLIN_000000200.5;Parent=transcript:GPLIN_000000200
pathogens_Gpal_scaffold_1 WormBase_imported CDS 47003 47013 . + 0 ID=cds:GPLIN_000000200;Parent=transcript:GPLIN_000000200
pathogens_Gpal_scaffold_1 WormBase_imported CDS 47446 47475 . + 1 ID=cds:GPLIN_000000200;Parent=transcript:GPLIN_000000200
pathogens_Gpal_scaffold_1 WormBase_imported CDS 47527 47655 . + 1 ID=cds:GPLIN_000000200;Parent=transcript:GPLIN_000000200
pathogens_Gpal_scaffold_1 WormBase_imported CDS 47711 47888 . + 1 ID=cds:GPLIN_000000200;Parent=transcript:GPLIN_000000200
pathogens_Gpal_scaffold_1 WormBase_imported CDS 47942 48028 . + 0 ID=cds:GPLIN_000000200;Parent=transcript:GPLIN_000000200
###
# Gene gene:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported gene 48851 67668 . - . ID=gene:GPLIN_000000300;Name=GPLIN_000000300;biotype=protein_coding
pathogens_Gpal_scaffold_1 WormBase_imported mRNA 48851 67668 . - . ID=transcript:GPLIN_000000300;Parent=gene:GPLIN_000000300;Name=GPLIN_000000300;info=method:InterPro accession:IPR001452 description:SH3 domain %0Amethod:InterPro accession:IPR002017 description:Spectrin repeat %0Amethod:InterPro accession:IPR002048 description:EF-hand domain %0Amethod:InterPro accession:IPR011992 description:EF-hand domain pair %0Amethod:InterPro accession:IPR013315 description:Spectrin alpha chain%2C SH3 domain %0Amethod:InterPro accession:IPR014837 description:EF-hand%2C Ca insensitive %0Amethod:InterPro accession:IPR018159 description:Spectrin/alpha-actinin %0Amethod:InterPro accession:IPR018247 description:EF-Hand 1%2C calcium-binding site
pathogens_Gpal_scaffold_1 WormBase_imported exon 67540 67668 . - . ID=exon:GPLIN_000000300.1;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 66788 66928 . - . ID=exon:GPLIN_000000300.2;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 66320 66394 . - . ID=exon:GPLIN_000000300.3;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 65702 65926 . - . ID=exon:GPLIN_000000300.4;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 65138 65239 . - . ID=exon:GPLIN_000000300.5;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 64951 65084 . - . ID=exon:GPLIN_000000300.6;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 64486 64675 . - . ID=exon:GPLIN_000000300.7;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 63771 63983 . - . ID=exon:GPLIN_000000300.8;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 63423 63504 . - . ID=exon:GPLIN_000000300.9;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 63059 63231 . - . ID=exon:GPLIN_000000300.10;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 62815 62997 . - . ID=exon:GPLIN_000000300.11;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 62428 62760 . - . ID=exon:GPLIN_000000300.12;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 62027 62096 . - . ID=exon:GPLIN_000000300.13;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 61825 61931 . - . ID=exon:GPLIN_000000300.14;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 61173 61397 . - . ID=exon:GPLIN_000000300.15;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 60831 60998 . - . ID=exon:GPLIN_000000300.16;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 60548 60676 . - . ID=exon:GPLIN_000000300.17;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 60396 60505 . - . ID=exon:GPLIN_000000300.18;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 59946 60017 . - . ID=exon:GPLIN_000000300.19;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 59768 59858 . - . ID=exon:GPLIN_000000300.20;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 59561 59647 . - . ID=exon:GPLIN_000000300.21;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 59294 59428 . - . ID=exon:GPLIN_000000300.22;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 58935 59069 . - . ID=exon:GPLIN_000000300.23;Parent=transcript:GPLIN_000000300
pathogens_Gpal_scaffold_1 WormBase_imported exon 58749 58892 . - . ID=exon:GPLIN_000000300.24;Parent=transcript:GPLIN_000000300
etc.
Sample 2 Protein fasta
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
>g1.t1
CWIQKIADGNEWKREPLVHFAKVVWLDLMRASLIASHCANVIYAGKQTDIDIDHRTIKLRLQEITAHLANWIAKMLEITWPTISRTRAKSAIGDSDIAES
KEAYQSVAEKIKEELGQFGAEKYDYAVIVFPNWTNPEAKAIICEANSCFELIDVKQINVIVVRIEDNETNFELACKANEWFTPRMESIMKNTIRMWWERQ
KELKLSDLANMLKEWLQFYRHLVLIHNWKFFSSAATITLGVTPTFVRSARAEKGFNHKIDPTFAEAEIFHVHMLL
>g2.t1
MAQIVNLISFLTLVISWPNALSSDTEAVKELATQFSSLTSSTWDIVNQLRFVSENAMEFVRAAGAVGAIIGRAVDIEFGPESDEQKAIKELHEFIKTKFE
ALQRHRQQESNAIKHNFIGTDYQNVLKI
>g3.t1
MKHNLVEKCQLPSQEQMERYAAVLELFQKIEQRHLLLLNQNDVILVEKYKSIKSSILAKSAVAPMQDKLNTILNSANSADEVWQALIKLNMDFHYAENTC
WIQSIADGNEWKREPLFHFAQLVCLDLMKMSLIALHCVNIMYAGKAIEHTLRQIELMLQEIASHMADWIAKMLEITWPTISRIYAKSAIGVSPIKESKDG
YQFVAEKIKEELVQFGAEKYDYAVIVFPNWTDPEAMAIISEANSYFELIDVKQINVIVVRIEDNETNFELACKAAEWFTPRIETKMKEAIKMWWDHKRHL
KLSDLADMLKEWLQFYRNLVLVHNWKFFSSAATITLGVTPTFVRSARAEKGFRHKIVPALFDAEIFHVHMLL
>g4.t1
MRAKCRKVRPKKDGDKMPKVMPKKGGDKMPTKWDKTPKNAGQKRMGQNAGKCCAAKKDGQMAKTKCGVGGVPILPREMGQNAGKPCPDDIRTTSVAIRRT
AHGHCMSFDVRDEFFDQSGVGIRFFVLILPRNGTNMMRMETLRETVEVDELIQNNLTIYNRITQRDEPFSRICRRFCTINEPVRLFYDGFKDHQRRVAKH
QQPSHNVQLDYPTSTVFGQRINIQPNFFGIHFANTTSSSSSEASSSNRFTNMDSVEMVVLLYRAERIGGWKDDDISDYEMSISNYFKDKYIGQHIRVLTI
STSYVQVEVDRAGNIIRSYFGFGLAVMILCSLFSNSLSAFFMHQFSIYKLPVAVFACLCPFMASGTALGILFFAGVRHSSILAVTPFLILALGVDDAFLM
IHSWQLASKRRRHEGGRAVMAHDALSVNLAVVDQLAEVLEDTGPAILISTLTNIFADLVGAFLGSEEITLLCIGNIASIIVDFFYQITLFTAVLVICARR
ELISARKRDNNKIHATNSNNLSEDDVKPTKSGGFRHSLERKFNAFKDIYICLISSLLVCIVWAAFLVVCLNALLKFEINLTLKKLFPPDSPLLEIEKYRE
EKVLPFYTTAQLFVNNPGDLSNATRRAHLNGLIDEMEHLKYAYPAESSLYFVRTYEEFLHGDSAGILDAAAEEENAKNGTATNEFDLSELETFLNWPEFR
HWRGLLKYHHTTHPQQEQSAESQGNGTELTLATNRSDGTQLDAMMTTVAYHGDELQDWYNRALMLREWRQVVDKYVPEFNVSVLHDDGLYLDLLETWYGR
GRMGKTIWYGRGEWGNATWYGRGNGDRENGTPINIWQSVVVTLLCMAAICGLFMCNMFVVFVTTCVIASIFIETLGIMALMDMTMDPVVMAAVTISIGFS
VDMPAHVSYHFHAAKWAGTLGGQNVHGIADQSVEHKIRHAFTSVGFPALQASICTCICVLSLAFVKIYTAQVFVRVLCVCIVLCAVHSLFLLPALFTFLD
AICKLFRQFMCQRR
>g5.t1
MNDNDRAISNHRQKQQQQQGGDINGAIVINNFTDQQHQHQQQQRQLINSAIHRRHRRHPRPRHPPLPTVTRRPNRFPPAAPSVSMRSLRVPIAAGPILWW
HIRTKLCSYASS
>g6.t1
MSELYLALQGEQCYYTVPSIHKGFNVKNLPFECPSFSIALEQQLQQIVDFSGCVKAIETWKAEHMYNRTDCLDSVKKAILESIIDAYNIYQNEADRMHRE
NVLHKYSHSIADLCAFVRTWQQRLNHLLGLLQKSDGLCGLEMMNVLFGQWTFQCCINDWRYRTIETALLAGSSAMHQLLVSWLLDAKLAPAALTRWFIRP
KFVPSSDDPNESQLQRPRHSFNNSVLSAAQLTMNGSANALNMSIAANTTTNYQNELCLFEAPPPTADAVIMASATTMTRNSASFFGTTTAGSDAASIGTG
TGTFVVAAAGAVVEDRIGWASQQQLILSVQLDTDVLPAPFYGCEELLNMIMDIGRKVHFLKMLRPAHANNEQQQSLCAQLPTNVWCTPSAIGRMATILRQ
MHAVVSEEMGRELMGRQRLMDHLCLVHGYFFMLRGDFASALEHILFIKTAKTTATKKCATPFSSRKNVSHSTKIAEKCVNDALAMCSPPANVADLMHLLC
GVKLTKREQMKCKIFSEKFMLKSRLREGQRNEFQAVAEFFSTNALRAYKRVFGFVSNLQLQRAQLDYTGQNLLRVVRQRLATRSEYRSSFYSMLIILQKV
RMFLNRLAHYVFEEVLLLESIYIFDKVRATKDPHVLLAGLYPLLSGVLLALFLPSALSKLHESVQQCIRNASDFVGSISKFCVFIEHSDSDVLDALLLAS
TDDDDDGTATNDDRDTNKNISNQHQQDDEDGTDHRHDVLSNGHPHHQQNCNANNDDCCDGIEQRENQPPPSEEEEEGQQQRAQQQQQQAQKVVTYNNTNS
NDNSSANGLRSAAENRNNSRTTDADQQQFHSVVFAISDSNVNFEASVAKLRNDLATLLAECSSSETKSRYFTPYWSKLNALLFHD
>g7.t1
MPWRTFESYTANDQNAQSFLVSLTEFLSPYAEIDQDKAGVSPEELRAAFFRCGGVNAHSWRILVLAAEGDNETTKFIGKLLDFLTAEGTSAPLTNCPPAA
APPNSQLANPPLQHQAGTSQASQFPFSAAIHSQSGLTCRQEVVGGIAGLKTPPDHIYDTISSSSSADDSHSPHKKLSTLPGHIYHTVMTTTSDGSQCSSV
FYDVPPPSSTIPRESVTTTPPPVPQHTSVVRPMSTSSTDGSQRSSVIYDVPPTPPPPVPQHTHGGEKNRKKNETPSPASKCSDAKKKRKETPKKDSDGKN
>g8.t1
MSKRRLRQELQDMVKNPLPEGCTARPLDQRTMNSWTVTIRGRAGTPYANGNFQLSMAFPRDYPFKAPQVTFTTRIFHPNIDTRGKVCLDILQTHWAPTLT
IEKVVISIIVLLNDPNPDNSPINAQAASLYRTDRAEFNRRAREMTLAYAR
>g9.t1
MNSFLFQPKAGAAVPTPQTSAAGGPPGAAEAPLSVKSSEKSGGTKKGPTPNASTMSRSLTYKPHVVSKDLALLQSEPNYGTLAGLDKEIFAKEQQAKGGM
PPPPMGAPQPQQQAPGPAMTPTTDPNYWTLKNLDADIFTKNAPAKPAAAPKAGPAPHGGGGQALTPTTDPNYGTLQNLDAEIFVKK
>g10.t1
MDQLPVTTTEITAAMANDQPLQTVLQFLRNRWPAKSTDSEISPYFPKRDKIFEVDNFLLYEDRVIVPTSLRSRILKALHIAHPGIVRMKELARRHVYWPG
MDSAIEKVVSTCEECQLGQKKPTKASLSPWPTTDKVFQRVHVDFAGPCSDGRQYLILIDSFSKWPEVYRMNTISAYATVFVLRSIIFQLGIPEEIVSDNG
TQFRSAEFATFCKEFGIKHIFTPPFHPQSNGQVERFVDTFKRSMKKMSKGEKDWIEKMLFAYRTTPHVALDGFSPDQLFFSRKLRTKLTLIHPKGEKAEN
FDQNSFKKGQSKYTDKMAQNFDRKHGSKLTNFFPNDHVLLINYRNNKTEWLQGKIVERLHNSLFHLNKICLSQPIPRW
>g11.t1
MYRHNLASAHNFLLCLILIVSAAYYDSSATAAHTIIISVLHHTSSSSVYWGQSAWKFCYAFGAHIDHRKGGFDRVLDLDYIIRAHQAVPNGVRYFANGRL
IKVFSAHAMLIRATPQQH
>g12.t1
MLGLRIPRNSPLLFNCCVSPPKSEFARFHVARSMTSGPSNFRHAFDHLIKSQQRRHYASGAMPKYTYNNRKADAILPDKWGAIFFLACSQSR
>g13.t1
MLGLRIPRNSSLLFNCCVSPPKSEFARFHVARSMISGPSNFRHAFDHLIKSQQRRHYASGAMPKYTYNNRKADALLPDKWGAIVFFGFFGLFIGYPIYGD
etc.
Sample 2 genome gff
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
000080K AUGUSTUS gene 1 3549 0.8 + . ID=g1;
000080K AUGUSTUS mRNA 1 3549 0.8 + . ID=g1.t1;Parent=g1
000080K AUGUSTUS intron 1 355 0.8 + . Parent=g1.t1;
000080K AUGUSTUS CDS 356 449 0.95 + 1 ID=g1.t1.CDS1;Parent=g1.t1
000080K AUGUSTUS exon 356 449 . + . ID=g1.t1.exon1;Parent=g1.t1;
000080K AUGUSTUS intron 450 1162 1 + . Parent=g1.t1;
000080K AUGUSTUS CDS 1163 1297 1 + 0 ID=g1.t1.CDS2;Parent=g1.t1
000080K AUGUSTUS exon 1163 1297 . + . ID=g1.t1.exon2;Parent=g1.t1;
000080K AUGUSTUS intron 1298 2313 1 + . Parent=g1.t1;
000080K AUGUSTUS CDS 2314 2450 1 + 0 ID=g1.t1.CDS3;Parent=g1.t1
000080K AUGUSTUS exon 2314 2450 . + . ID=g1.t1.exon3;Parent=g1.t1;
000080K AUGUSTUS intron 2451 2838 1 + . Parent=g1.t1;
000080K AUGUSTUS CDS 2839 3054 1 + 1 ID=g1.t1.CDS4;Parent=g1.t1
000080K AUGUSTUS exon 2839 3054 . + . ID=g1.t1.exon4;Parent=g1.t1;
000080K AUGUSTUS intron 3055 3248 1 + . Parent=g1.t1;
000080K AUGUSTUS CDS 3249 3385 1 + 1 ID=g1.t1.CDS5;Parent=g1.t1
000080K AUGUSTUS exon 3249 3385 . + . ID=g1.t1.exon5;Parent=g1.t1;
000080K AUGUSTUS intron 3386 3439 1 + . Parent=g1.t1;
000080K AUGUSTUS CDS 3440 3549 1 + 2 ID=g1.t1.CDS6;Parent=g1.t1
000080K AUGUSTUS exon 3440 3549 . + . ID=g1.t1.exon6;Parent=g1.t1;
000080K AUGUSTUS stop_codon 3547 3549 . + 0 Parent=g1.t1;
000080K AUGUSTUS gene 9538 11185 0.39 + . ID=g2;
000080K AUGUSTUS mRNA 9538 11185 0.39 + . ID=g2.t1;Parent=g2
000080K AUGUSTUS start_codon 9538 9540 . + 0 Parent=g2.t1;
000080K AUGUSTUS CDS 9538 9558 0.98 + 0 ID=g2.t1.CDS1;Parent=g2.t1
000080K AUGUSTUS exon 9538 9558 . + . ID=g2.t1.exon1;Parent=g2.t1;
000080K AUGUSTUS intron 9559 9604 0.99 + . Parent=g2.t1;
000080K AUGUSTUS CDS 9605 9653 0.98 + 0 ID=g2.t1.CDS2;Parent=g2.t1
000080K AUGUSTUS exon 9605 9653 . + . ID=g2.t1.exon2;Parent=g2.t1;
000080K AUGUSTUS intron 9654 10139 0.99 + . Parent=g2.t1;
000080K AUGUSTUS CDS 10140 10306 0.97 + 2 ID=g2.t1.CDS3;Parent=g2.t1
000080K AUGUSTUS exon 10140 10306 . + . ID=g2.t1.exon3;Parent=g2.t1;
000080K AUGUSTUS intron 10307 11035 0.85 + . Parent=g2.t1;
000080K AUGUSTUS CDS 11036 11185 0.39 + 0 ID=g2.t1.CDS4;Parent=g2.t1
000080K AUGUSTUS exon 11036 11185 . + . ID=g2.t1.exon4;Parent=g2.t1;
000080K AUGUSTUS stop_codon 11183 11185 . + 0 Parent=g2.t1;
000080K AUGUSTUS gene 13004 16505 0.55 + . ID=g3;
000080K AUGUSTUS mRNA 13004 16505 0.55 + . ID=g3.t1;Parent=g3
000080K AUGUSTUS start_codon 13004 13006 . + 0 Parent=g3.t1;
000080K AUGUSTUS CDS 13004 13166 0.55 + 0 ID=g3.t1.CDS1;Parent=g3.t1
000080K AUGUSTUS exon 13004 13166 . + . ID=g3.t1.exon1;Parent=g3.t1;
000080K AUGUSTUS intron 13167 13250 0.93 + . Parent=g3.t1;
000080K AUGUSTUS CDS 13251 13301 0.93 + 2 ID=g3.t1.CDS2;Parent=g3.t1
000080K AUGUSTUS exon 13251 13301 . + . ID=g3.t1.exon2;Parent=g3.t1;
000080K AUGUSTUS intron 13302 13442 0.93 + . Parent=g3.t1;
000080K AUGUSTUS CDS 13443 13524 0.94 + 2 ID=g3.t1.CDS3;Parent=g3.t1
000080K AUGUSTUS exon 13443 13524 . + . ID=g3.t1.exon3;Parent=g3.t1;
000080K AUGUSTUS intron 13525 13784 0.96 + . Parent=g3.t1;
000080K AUGUSTUS CDS 13785 13878 0.99 + 1 ID=g3.t1.CDS4;Parent=g3.t1
000080K AUGUSTUS exon 13785 13878 . + . ID=g3.t1.exon4;Parent=g3.t1;
000080K AUGUSTUS intron 13879 13943 1 + . Parent=g3.t1;
000080K AUGUSTUS CDS 13944 14072 1 + 0 ID=g3.t1.CDS5;Parent=g3.t1
000080K AUGUSTUS exon 13944 14072 . + . ID=g3.t1.exon5;Parent=g3.t1;
000080K AUGUSTUS intron 14073 15000 1 + . Parent=g3.t1;
000080K AUGUSTUS CDS 15001 15137 1 + 0 ID=g3.t1.CDS6;Parent=g3.t1
000080K AUGUSTUS exon 15001 15137 . + . ID=g3.t1.exon6;Parent=g3.t1;
000080K AUGUSTUS intron 15138 15394 1 + . Parent=g3.t1;
000080K AUGUSTUS CDS 15395 15610 1 + 1 ID=g3.t1.CDS7;Parent=g3.t1
000080K AUGUSTUS exon 15395 15610 . + . ID=g3.t1.exon7;Parent=g3.t1;
etc.
The goal is to obtain operon.db and genome.db files that follow this format
Because gff and protein names are so variable between species, it is hard to standardize a script to deal with such huge variation in formatting and naming schemes. However, the file format below is what we are aiming for. Notice that the typical +/- designation for strand is now D/C which stands for Direct and Complementary strand. Relative gene number is essentially providing a count for opscan, so just number as integers from 1- onward.
1
2
3
4
><genename@Species@Scaffold> <relative gene number> <start> <end> <strand in C and D format, not - and +> <scaffold_name>
MADQQQQQQDDLGPPPDPVMEVPPPKEIKVLESADDIQSRRTEVLDHYAQFQGFATIKRERLEEARQFQYFKRDADELEIWILEKLQTAAEESFRDPTNLQAKIQKHEAFVAEVQAHSNAITKLDKTGNDMIQHDHYEKDTIRKRLDRLHELWDRLFAMLEGKGIKLQQTLKLLQFVRKCDEMLYWIRDKIQFVSSDDLGADLEHVEVMQRKFDEFLKELKSHESRVLDINHEANTLIDEGHPEQQQIHGKRDEVNEAWHKLGTLTSTRGEALFGAQQIQRFYRDIDETLAWMGEKELTLATDDYGRDLNNVQALQRKHEGTERDLAALESKMDKLGSETNRLCQLYPEKSGDIATKMDEAKGRWEALKLGAEQRKRGLDRSYNRHRFMADYRELGEWIKGIQALIQSSELAKDVAGAEALLEQHQEHKGEIEARADSFKQTAETGQRLLDEEDIDNAXXXXXXXXXXXXXXXXXXXXXXNLISFLATGAKLQEANQEQLFNRNIEDVELWLSELEGQLASEDFGKDLVSVQNLQKKLGLVESDYNAHQERIDAIGHQTREFHDTGHFNAPLISKKFDVLQQRFEALREPLQRRKQKLAESLLGHQLFRDIDHELAWIREKEQIAASTNRGRDLVGVQNLIKKQNALHTEIANHEQQMEQVAQSAEQMVQRGHFLAPDIRDKKSQLRDNWRALKTKADKRKQDLQDSLQAHQYLADANEAESWMRDKEPLVGSTDYGKDEDSAESLLKKHRALMSDLEAFNTTIEELRKEAGQCKYQEQLGGQLGKECVVALYEYTEKSPREVSMKKGDVLTLLNASNKDWWKVEVSDRQGFVPAAYVKRIEPGSQHHHQAMAGQPMNTINAKQSQSEDQYKRLLMLGEQRKRKLEEACKAYQLLREANDLAEWIRSRETVASQQEIGQDLEQVEVLQNKFNDFKNDLKANEVRLQEMNQIATRLSQMGQTETAVRIRQQIDDLNARWKALEQKAEERGQQLESAHEVQRFHKDVDETKDWIQEKNETLDSEDFGRDLRSVQALQRKHEGVERDLAALGDKIRALDEKANRLRQTHPEAAEQIYDLQRQLNEQWTTLTQKANRRKDKLLDRPCVSMKKGDVLTLLNASNKDWWKVEVSDRQGFVPAAYVKRIEPGSQHHHQAMAGQPMNTINAKQSQSEDQYKRLLMLGEQRKRKLEEACKAYQLLREANDLAEWIRSRETVASQQEIGQDLEQVEVLQNKFNDFKNDLKANEVRLQEMNQIATRLSQMGQTETAVRIRQQIDDLNARWKALEQKAEERGQQLESAHEVQRFHKDVDETKDWIQEKNETLDSEDFGRDLRSVQALQRKHEGVERDLAALGDKIRALDEKANRLRQTHPEAAEQIYDLQRQLNEQWTTLTQKANRRKDKLLDSYGQCFLKAALIEKRSKLGESQTLQQFSRDAERWRIGWRRSSKSRRRRPTGPTNIQQKHQKQQAFEAELSANADRIATLISAGQNLITAAKCGGGETAVSARLRALNDQWEQLVKTTTEKSHRLKEANKQKTFMAAVKDLEFWLGEVETLLASEDYGRDLASIENLLKKHQLLEADITAHADRVEDMNRQADALLESEQFDQAQIDGRRRNINERYERVKDAANVRRDKLNKAITVHQFLRDIDEEESWIKEKKLLVSSDDYGRDLAGVQNLRRKHRRFDTELATHQPQVQVVRSKGMELMASSEIGVPEIVKRIKALDQSWTHMVELTEDRHKKLQESEEFQNFIGRVEEEEAWLNEKQQILSSPNVGENMAAVQGLLKKHDTFEVDLQMHQQRIDELQRQGEELINAGNHHAKNIGTRMEQLVKHLILVKELAVRRLQKLRDNNAYMQFMWKCDVVESWIGDKEPHVRSTDFGRDLSSVQLLLNKQDTFDNGLNNFEHEGIQRVTELKDQLLEAEHEQSAAIEQRHQAVIQRWQQLLMNSLERRKKLTEAQAHFKNIEDLYLTFAKKASAFNSWFENAEEDLTDPVRCNSLEEIREHSTVGLAQAWDQLDQLAMRMQHNLEQQIQARNQSGVSEEALREFSMMFRHFDREKLGRLDHQQFKSCLRALGYDLPMVDEGQPEPEFQRILDLVDPNRDGYVTLQEFMAFMINKETENVRSSEEIEMAFRALSKEFRPYVIAEELFANLTPEQADYCINRMKPYVDAASGRTIAGALDFEQFVQSVFMN
><genename@Species@Scaffold> <relative gene number> <start> <end> <strand in C and D format, not - and +> <scaffold_name>
VGSAGLLCETGGFCPWLRAVSLRGGAWTSGESCLWSASAACMLVRAGVRQESGLSSHARKLHSSEKEARANLHAPRCLSAMPKFTATFEANTLSELLSQIKGVLAEGNSVGVGAKKPSVGEAGGVAKAGGKAAKRKRLRQQAAAAVVAAKHAKGGDEEAAETSEKQVDPSSTKPPKKDKPHRKSLEGKE
sample 1 operon.db file example
1
2
3
4
5
6
7
8
9
10
11
12
13
14
>GPLIN_000000100@G.pallida@1 1 1060 32 D 1
VGSAGLLCETGGFCPWLRAVSLRGGAWTSGESCLWSASAACMLVRAGVRQESGLSSHARKLHSSEKEARANLHAPRCLSAMPKFTATFEANTLSELLSQIKGVLAEGNSVGVGAKKPSVGEAGGVAKAGGKAAKRKRLRQQAAAAVVAAKHAKGGDEEAAETSEKQVDPSSTKPPKKDKPHRKSLEGKE
>GPLIN_000000200@G.pallida@2 2 48028 47003 D 1
MSKAAEPLLKDAIISLGPPNSTIVCAVSGHPTSFPQRLCNAIFSKIYELGITILVSKLEGRFLHVQLGSGMDALMALSMDGVVLGEDLRLEVCLGFAGCWLDLLMADLQCLGVVREIRMRKVMNCLAMGKRLFWTMFRSLISPM
>GPLIN_000000300@G.pallida@3 3 67668 48851 C 1
MADQQQQQQDDLGPPPDPVMEVPPPKEIKVLESADDIQSRRTEVLDHYAQFQGFATIKRERLEEARQFQYFKRDADELEIWILEKLQTAAEESFRDPTNLQAKIQKHEAFVAEVQAHSNAITKLDKTGNDMIQHDHYEKDTIRKRLDRLHELWDRLFAMLEGKGIKLQQTLKLLQFVRKCDEMLYWIRDKIQFVSSDDLGADLEHVEVMQRKFDEFLKELKSHESRVLDINHEANTLIDEGHPEQQQIHGKRDEVNEAWHKLGTLTSTRGEALFGAQQIQRFYRDIDETLAWMGEKELTLATDDYGRDLNNVQALQRKHEGTERDLAALESKMDKLGSETNRLCQLYPEKSGDIATKMDEAKGRWEALKLGAEQRKRGLDRSYNRHRFMADYRELGEWIKGIQALIQSSELAKDVAGAEALLEQHQEHKGEIEARADSFKQTAETGQRLLDEEDIDNAXXXXXXXXXXXXXXXXXXXXXXNLISFLATGAKLQEANQEQLFNRNIEDVELWLSELEGQLASEDFGKDLVSVQNLQKKLGLVESDYNAHQERIDAIGHQTREFHDTGHFNAPLISKKFDVLQQRFEALREPLQRRKQKLAESLLGHQLFRDIDHELAWIREKEQIAASTNRGRDLVGVQNLIKKQNALHTEIANHEQQMEQVAQSAEQMVQRGHFLAPDIRDKKSQLRDNWRALKTKADKRKQDLQDSLQAHQYLADANEAESWMRDKEPLVGSTDYGKDEDSAESLLKKHRALMSDLEAFNTTIEELRKEAGQCKYQEQLGGQLGKECVVALYEYTEKSPREVSMKKGDVLTLLNASNKDWWKVEVSDRQGFVPAAYVKRIEPGSQHHHQAMAGQPMNTINAKQSQSEDQYKRLLMLGEQRKRKLEEACKAYQLLREANDLAEWIRSRETVASQQEIGQDLEQVEVLQNKFNDFKNDLKANEVRLQEMNQIATRLSQMGQTETAVRIRQQIDDLNARWKALEQKAEERGQQLESAHEVQRFHKDVDETKDWIQEKNETLDSEDFGRDLRSVQALQRKHEGVERDLAALGDKIRALDEKANRLRQTHPEAAEQIYDLQRQLNEQWTTLTQKANRRKDKLLDRPCVSMKKGDVLTLLNASNKDWWKVEVSDRQGFVPAAYVKRIEPGSQHHHQAMAGQPMNTINAKQSQSEDQYKRLLMLGEQRKRKLEEACKAYQLLREANDLAEWIRSRETVASQQEIGQDLEQVEVLQNKFNDFKNDLKANEVRLQEMNQIATRLSQMGQTETAVRIRQQIDDLNARWKALEQKAEERGQQLESAHEVQRFHKDVDETKDWIQEKNETLDSEDFGRDLRSVQALQRKHEGVERDLAALGDKIRALDEKANRLRQTHPEAAEQIYDLQRQLNEQWTTLTQKANRRKDKLLDSYGQCFLKAALIEKRSKLGESQTLQQFSRDAERWRIGWRRSSKSRRRRPTGPTNIQQKHQKQQAFEAELSANADRIATLISAGQNLITAAKCGGGETAVSARLRALNDQWEQLVKTTTEKSHRLKEANKQKTFMAAVKDLEFWLGEVETLLASEDYGRDLASIENLLKKHQLLEADITAHADRVEDMNRQADALLESEQFDQAQIDGRRRNINERYERVKDAANVRRDKLNKAITVHQFLRDIDEEESWIKEKKLLVSSDDYGRDLAGVQNLRRKHRRFDTELATHQPQVQVVRSKGMELMASSEIGVPEIVKRIKALDQSWTHMVELTEDRHKKLQESEEFQNFIGRVEEEEAWLNEKQQILSSPNVGENMAAVQGLLKKHDTFEVDLQMHQQRIDELQRQGEELINAGNHHAKNIGTRMEQLVKHLILVKELAVRRLQKLRDNNAYMQFMWKCDVVESWIGDKEPHVRSTDFGRDLSSVQLLLNKQDTFDNGLNNFEHEGIQRVTELKDQLLEAEHEQSAAIEQRHQAVIQRWQQLLMNSLERRKKLTEAQAHFKNIEDLYLTFAKKASAFNSWFENAEEDLTDPVRCNSLEEIREHSTVGLAQAWDQLDQLAMRMQHNLEQQIQARNQSGVSEEALREFSMMFRHFDREKLGRLDHQQFKSCLRALGYDLPMVDEGQPEPEFQRILDLVDPNRDGYVTLQEFMAFMINKETENVRSSEEIEMAFRALSKEFRPYVIAEELFANLTPEQADYCINRMKPYVDAASGRTIAGALDFEQFVQSVFMN
>GPLIN_000000400@G.pallida@4 4 74810 70588 C 1
MITTRLSTCHCTFVGGFFRSHRSENFKKINKAVNIKCQPRMPKKLLPVTSELKTHFEILLERFLDQKYSQAYLNLFTLYLKTFKQFTAFSEEQAIEALAKLEDFFTKMSLDSEIQLSDLNAQLLTLLKNYDSDERHSWEVSELEQLALYTKHITGNVSDWIQNELEDAWPDELSRAAKNALGIEQISDPYAYNKTAHIIKDAVSSRGVPGFIYTILVSLDWDPIANFWYKCNPDYCFPVKGFHGINFVVLRHEIGSQQRAEQAEKWFEAQKSAMIKTIRDNDKISDLGSLIRKGAKVRRKLEEFAH
>GPLIN_000000500@G.pallida@5 5 78993 78641 D 1
MDERFEQSLEGCWFFETAKRMASDFNQVSECLETQFEVREWESEFSGEEDAPLPKMREICSRANLHLMAQDEPSGGKQKSAEVVAAIEDNCF
>GPLIN_000000600@G.pallida@6 6 85516 82522 D 1
MLINIGFFFVSIKLLFLVNIDKVSSCALTRYDKGGCCKRNVWSEAIVCPRPYEPNCNIFEYNCVGCTYVFDKANACSVRVYCSCFYCCDPKYAESCPDGCRRCILDGIGGCPENVPDINSSSSMPESEAKAWKHFNLIDVDKSGSLSLNEAVEHLGSKPDNGTFAKHLAKNLSWFAEIDRNANNQIEPWEFDRSLIGVNNHTNG
>GPLIN_000000700@G.pallida@7 7 95835 94711 D 1
etc.
sample 2 genome.db files example
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
>g1@H.glycines@000085K 1 6346 3036 C 000085K
MSSVRSISSTARAGNVLVLLDLYASGFASSNTSPTIPMLTSRTISFPLNFWLAMLLVIASISGTMLIILLMNAFAHRLDITVHKNGLAVIIEAFLEWLYGFNNFRVAFLFMLLHDAPITLANFFLLGACRCAGPNVLSWPLLLSALAIALSLCWRLLLLRFAYARLVFRRPSPDPNSAFPSRSHSFRQQFAADQCTASQNEVNSQLDELWAIRRARAIVYGEKRGKCWEGTAGETLYLGEEEEGARGTGRSMGQCCGFWSSWVLLKGFDLFCAFPNSFSHRHKCAKTFVRHFSLLFHYVIFGFSLCFSALLILLNVVLIFSTQLVGPNQWPPEISRLCISVSPHRRIVRPLFLPQSSLFSSFLLPPKQMNNGTAADLRSHSLVQCKPIWDLPHQQTLVDKWAPGLSRKSPSPWQSRFPLQDGRVLAVSTHFVSDHSEEQSPCHTLFYDFALLTVGADNSSKTTKCARQRSSGWFFANARNVLAFPYFWACSPSISIRKGTLINCQSLTKH
>g2@H.glycines@000085K 2 9212 7856 C 000085K
MNAFCSAPSEGPVTIELRQKIVDFFKPEHLEVECESRLHNVPKGAEKHFRVQIVSDKFNGMTQLHRQRMVNKLLAEELREKIHALRIEAKIPSEWHGTKQTPAPPCEGGEKKRRQSEQPKRQ
>g3@H.glycines@000085K 3 14129 11906 C 000085K
MLSPLASSSSARPSYSRHHKQSTYFHRPTLIKSEPEELCSSVPPSLVPKQLRPPPLIDTSTPESLERDFHAWLKGELSRSERPFCARHSDCFSAQSPPPSAMFLTPLGNVKNELVVCPTDFGGDFVFGMELEPSNSDFCASTAAFAPSISMNGPFPPMQTQMIRMPMPSLSASITQEDQQMAYLLSSAGQSPSGSSQHSSLEGSPLQPNCHQTMLLSIGKYELNDRKMLTELSGEVKTDGAKKMAATDAAMTAPNSPGGRSSGTNQLGRTYSPGLPLSMVERQKIVQLFHEGWKICDISKYLCVTHSCVSKILQRFRATGSVRPKDAKEGRQESPLVAAIRDYRQRLGIMRQSEIREQLIRDGLCRRENAPSRSSINHILRTKLSGLEAPTIGKRSSNQTTENANGKRQTDGRGENDGTAANAATARVYYDE
>g4@H.glycines@000085K 4 20477 18394 C 000085K
MVPFRFRLHLPLAIRSADCSIIRSLHAHRHFCERMIPLKSSSASVTVSTSSAMFVGADSCRRRSVRRFSAPFPKMLPTALLLCASIVFMTNPSVDAEISIKIERHFPCSAASGPKKENLIIKFPSYKSTGVAFKEEKDDAGHKCFQMGGGKVEVYAPGLDGNKKYYVHVETRIGIHGKPERCVNPDKEGCGGIGSCVHCDICQTIGGFRQFIQIFKGDKPAECSAKGLPPGTYDNLSLKVCLPSKDELLPFLDQNSSRAEQLWDVFVSSRARSGGEIPLVIAARIFDRPINKLSIKELNDALHNTKKGMIGCHWIYATVRSINQD
>g5@H.glycines@000085K 5 27425 26288 D 000085K
MDIPPFIFLLFLVPTLSVQLTIDDEEFGLNLQLIKHQPCPFKSDKSKWDRKLEFEAGNEQEGPKLVQRPNEPNCYSIGGKVTVFSEFKGEFSIYLELRSSANKKQVPEPCHNRKDDGCGGFGSCLYCDACQTLEENRGLKAQLLVDGKSISCGQELSPGTYENMELVFCLPNAEEMLKSQGLTKETFKSLIQTEDGTNLRSLGVFATIYVFDRDVRKLVQTQQKVETVYRKTKRTLFKDEPLPPDVYWSLPFNSVIKEQKMFVACHKIFGNMQIRSNK
>g6@H.glycines@000085K 6 30496 27473 C 000085K
MLNPSVNKIHFSKDELDKILRDADEQRDFPALLRSLGQLAFRPTDGFSLPSSALSQMASHFASLSPFSAFHWANGLFALLPSAENHANSEVLIVSLLRSLVDNVPLLNANKLRAFVKTEISRQCVSFWQIGKAHLASALTLCVLRILLMELSYLEAIQNGVCRADLIAEAQNIWSQMLSADELLKVDPGAFLRLSSSLFHSALGLDYPLTNSLEANIVVCLKKCSNEMGSQNGCDSLYCASLFDLLSAHLATFCQILPGDVCRAMFEFLIAHSERDNLEILLKQLAKNSAIRCGECCEKVFRSLYDLPMPSPSFNLEEIWSIETKISGKKETQAQKIDESQLSWASLLSLSDECLANSLVGVSNAKIESIVLSADFAVFQPEICLRLLISLMKIKRTLVIPFRCEFVHLILAEEAPKSRSLEFGAKLLEMHAHSADSDAFPAKSDFVSFCLAIFQPIDQLDCDQCAKRDFLLAELYFLRALFSVLPHLLTSFAPLIVQLLPDFFVSSNFYRRTLREWPESRFADRLEHSLALLCREIVRRKKCFNKLLPFVISSSLTGDRELCFPMYLLFSALDENDLALLSSNLSAAEKLKFSRLKVQFYASNKIIG
>g7@H.glycines@000085K 7 32733 30713 C 000085K
MNLTRVNSNDHPELYQLIGPIEELYQLIPIGQIDVASDYVNYQLIGPIHAINDAVFYNTLALSNQLEAPIQLLAIEGPPQFQLVGPVPRLARIRCRFANDSHRLVDLVWTDNNHRMYRYAKLAHTDFIDITTFEGHQWVFCDALDGELNSLNPGGVKKFIASIHQNSHRRRLRVSAISSVEGLTLKRVILRLLASKIFDHWNSTDCCDCRSIGLFLETLRLPVCIQVQLANFLISCILYQFKFDSNRSNIGQKHLFDGLVNFIRSKTQIYYEN
etc.
#Code to convert Sample 1 to operon.db
1
2
#essentially this approach assumes the same naming scheme for genes and proteins, puts each file into single line format, sorts, and pastes together.
paste <( awk '$3=="gene" {print $1,$4,$5,$7,$9}' globodera_pallida.PRJEB123.WBPS7.annotations.gff3 |sed 's/ID=gene://g' |sed 's/;/\t/g' |sed 's/pathogens_Gpal_scaffold_//g' |awk '{print $5,NR,$3,$2,$4,$1}' |less) <(awk '{print $1}' globodera_pallida.PRJEB123.WBPS7.protein.fa |tr "\n" "\t" |tr ">" "\n" |sed 's/\t/#/1' |sed 's/\t//g' |sed 's/#/\t/g' |sort -k1,1V |sed '/^$/d') |awk '{print ">"$1"@G.pallida@"$6,$2,$3,$4,$5,$6"\n"$8}' |sed 's/-/C/g' |sed 's/+/D/g' >operon.db
#Code to convert sample 2 to genome.db
1
2
#This uses the same approach, but the protein file includes proteins predicted from alternatively spliced transcripts. Since primary transcripts are of the most importance and we want to prevent artificial duplication, we will grep "t1" when the fasta file is single lines.
paste <(awk '$3=="gene" {print $1,$4,$5,$7,$9}' 738.polished.mitofixed.repmod.gff3 |sed 's/ID=//g' |sed 's/;/\t/g' |awk '{print $5,NR,$3,$2,$4,$1}' ) <(awk '{print $1}' 738.polished.mitofixed.repmod.aa |tr "\n" "\t" |tr ">" "\n" |sed 's/\t/#/1' |sed 's/\t//g' |sed 's/#/\t/g' |grep "t1" |sort -k1,1V ) |awk '{print ">"$1"@H.glycines@"$6,$2,$3,$4,$5,$6"\n"$8}' |sed '/^$/d' |sed 's/\.t1//g' |sed 's/-/C/g' |sed 's/+/D/g' >genome.db
Beware, the code above assumes that the genes are sorted correctly, yours may be different.
Run opscan
1
2
#-S no classes computed, -b two or more operon.db genes can be a homolog with a genome.db gene, -C output suboperons in operon genes
./opscan -S -b -C -Q -O operon.db genome.db >iadhorePlazaTable
Sample of the output above
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
CO List of similar genes with operon GPLIN_000000100:
CO gene g12170 (score 31.85 dlen ratio 1.17)
CO gene g27614 (score 31.85 dlen ratio 1.08)
CO gene g22305 (score 31.21 dlen ratio 1.06)
CO gene g9207 (score 29.20 dlen ratio 1.02)
CO gene g25628 (score 28.25 dlen ratio 1.21)
CO gene g13794 (score 28.05 dlen ratio 1.27)
CO List of similar genes with operon GPLIN_000000200:
CO gene g11691 (score 32.56 dlen ratio 1.27)
CO gene g24814 (score 30.41 dlen ratio 1.08)
CO gene g15389 (score 29.84 dlen ratio 1.11)
CO gene g15392 (score 29.38 dlen ratio 1.11)
CO gene g10490 (score 28.17 dlen ratio 1.25)
CO gene g10488 (score 28.17 dlen ratio 1.25)
CO List of similar genes with operon GPLIN_000000300:
CO gene g16041 (score 70.59 dlen ratio 1.11)
CO gene g6503 (score 30.20 dlen ratio 1.08)
CO gene g25240 (score 29.64 dlen ratio 1.07)
CO gene g14467 (score 26.47 dlen ratio 1.10)
CO gene g13713 (score 24.66 dlen ratio 1.23)
CO gene g7023 (score 24.66 dlen ratio 1.23)
CO List of similar genes with operon GPLIN_000000400:
CO gene g29333 (score 38.45 dlen ratio 1.24)
CO gene g25693 (score 29.49 dlen ratio 1.30)
CO gene g25617 (score 29.02 dlen ratio 1.29)
CO gene g18315 (score 28.36 dlen ratio 1.27)
CO gene g11308 (score 27.85 dlen ratio 1.19)
CO gene g3825 (score 27.27 dlen ratio 1.08)
CO List of similar genes with operon GPLIN_000000500:
CO gene g21617 (score 31.30 dlen ratio 1.23)
CO gene g28038 (score 30.65 dlen ratio 1.23)
CO gene g5356 (score 29.47 dlen ratio 1.21)
CO gene g6095 (score 28.57 dlen ratio 1.19)
CO gene g6092 (score 28.57 dlen ratio 1.19)
CO gene g27039 (score 28.49 dlen ratio 1.26)
CO List of similar genes with operon GPLIN_000000600:
CO gene g1861 (score 47.97 dlen ratio 1.15)
CO gene g25809 (score 47.68 dlen ratio 1.00)
CO gene g1868 (score 43.01 dlen ratio 1.10)
CO gene g25829 (score 41.27 dlen ratio 1.00)
CO gene g1862 (score 39.47 dlen ratio 1.07)
CO gene g11644 (score 33.72 dlen ratio 1.07)
CO List of similar genes with operon GPLIN_000000700:
CO gene g3508 (score 30.92 dlen ratio 1.22)
CO gene g25120 (score 29.57 dlen ratio 1.27)
CO gene g11695 (score 28.33 dlen ratio 1.24)
CO gene g18323 (score 28.17 dlen ratio 1.28)
CO gene g17973 (score 28.17 dlen ratio 1.28)
CO gene g23796 (score 27.60 dlen ratio 1.29)
Reformat opscan output to suitable format for iAdhore synteny
Because a greater number of syntenic regions in the genome may indicate when our ortholog calls are the most accurate, applying filtering criteria to the opscan output will be informative.
1
2
3
4
5
6
7
#reformatting the above will generate a one to one ortholog relationship of the top scoring gene :
less iadhorePlazaTable |cut -f 2- |sed 's/CO List of similar genes with operon //g' |sed 's/://g' |grep -A 1 "GPLIN" |sed 's/--//g' |sed '/^$/d' |awk '{if(NF>1) {print $2} else {print "#"$0}}'|tr "\n" "\t" |tr "#" "\n" |sed '/^$/d' |less
#However since this does not provide any cutoffs, there are probably inaccurately called orthologs with low scores or poor length ratios. This command filters the opscan output for a NWS score of 40 and a protein length ratio of .8>x<1.2.
less iadhorePlazaTable |cut -f 2- |sed 's/CO List of similar genes with operon //g' |sed 's/://g' |grep -A 1 "GPLIN" |sed 's/--//g' |sed '/^$/d' |awk '{if(NF>1 && $4>40 && $7>.8 && $7<1.2) {print $2} else if (NF==1){print "#"$0}}' |grep "g" -B 1 |sed 's/--//g' |sed '/^$/d' |tr "\n" " " |tr "#" "\n" |sed '/^$/d' |sed 's/@/\t/g' |awk '{print $1"\t"$4}' >OrthologousGenes.list
#sample of the output from the code above (tab delimited as is required for iadhore)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
GPLIN_000000100 g12170
GPLIN_000000200 g11691
GPLIN_000000300 g16041
GPLIN_000000400 g29333
GPLIN_000000500 g21617
GPLIN_000000600 g1861
GPLIN_000000700 g3508
GPLIN_000000800 g17807
GPLIN_000000900 g25809
GPLIN_000001000 g2058
GPLIN_000001100 g17530
GPLIN_000001200 g23779
GPLIN_000001300 g18358
GPLIN_000001400 g5028
GPLIN_000001500 g18095
GPLIN_000001600 g23645
GPLIN_000001700 g25395
GPLIN_000001800 g29162
GPLIN_000001900 g24049
GPLIN_000002000 g10142
GPLIN_000002100 g21109
GPLIN_000002200 g10141
GPLIN_000002300 g24950
GPLIN_000002400 g25562
GPLIN_000002500 g19960
GPLIN_000002600 g10139
GPLIN_000002700 g10574
GPLIN_000002800 g18246
GPLIN_000002900 g18237
GPLIN_000003000 g8285
GPLIN_000003100 g16829
GPLIN_000003200 g28534
GPLIN_000003300 g3532
GPLIN_000003400 g14435
GPLIN_000003500 g19032
GPLIN_000003600 g25157
GPLIN_000003700 g13927
GPLIN_000003800 g4160
etc.
Infer Synteny from ortholgous GENES
First there are two types of synteny.
- Is using DNA to DNA alignments to identify conserved genomic fragments (Mummer, BLAST, etc).
- Is to use gene sequences only, to identify conserved gene order (iAdhore,mummer(promer)).
Both types of synteny are useful for inferring chromosomal rearrangements and for identifying highly conserved genes. The first type of synteny is only appropriate for very closely related organisms, as nucleotide similarity is quickly lost when species diverge. The second type of synteny is useful for finding conserved chromosome fragments in more divergent species. This allows for lots of change in intergenic regions, while we can rely more on evolutionary constrained genes to infer conserved gene order.
Iadhore is a program that uses gene order to identify synteny, and thus can be leveraged in many species comparisons.
First step here is to have a list of orthologs between your two targeted species already calculated. This can be done with blast, opscan, orthoMCL, etc. For this part of the tutorial, I will be using the prerequisite files (operon.db and genome.db) as well as the orthologs that were calculated above (OrthologousGenes.list)
Obtain prerequisite files
genome.db
1
2
3
4
5
6
7
8
9
10
11
>g1 1 1923 1 + 000085K
CQPAFNSTQHQHQCAPSSSTHFVATTHHFIMSDMSAYMGSGPAEPAPSSGAAEASPSSGVASAYMGSGPAEPAPSSGAAEASPSSGVASAYMGASPAYEPPAQEKSPDQSAYMFIIAVKNEEEHQQNRLRPYEIDPTRGIAERPRLKKYLRNDEAIRRLVIDFDHKENKNQADIINHIHALQYRLGMKGFEEWD
>g2 1 2912 2263 + 000085K
MGRGAHLPEDPQPRPRPCRWPTQAKLAGTSGFFVQLDELYKTVSMAAFRFAFPDGRSPPVPPLSVILRQSLLANHSKFRQEPEKLRHFYDNGETNAFMRSTFRDDQHSKKIVMGNGGGGEKEEKREDEKRK
>g3 1 6346 3036 - 000085K
MSSVRSISSTARAGNVLVLLDLYASGFASSNTSPTIPMLTSRTISFPLNFWLAMLLVIASISGTMLIILLMNAFAHRLDITVHKNGLAVIIEAFLEWLYGFNNFRVAFLFMLLHDAPITLANFFLLGACRCAGPNVLSWPLLLSALAIALSLCWRLLLLRFAYARLVFRRPSPDPNSAFPSRSHSFRQQFAADQCTASQNEVNSQLDELWAIRRARAIVYGEKRGKCWEGTAGETLYLGEEEEGARGTGRSMGQCCGFWSSWVLLKGFDLFCALCRRLLCLLLSLFLCSVCLSVGCVPCLHHYSCSPNSFSHRHKCAKTFVRHFSLLFHYVIFGFSLCFSALLILLNVVLIFSTQLVGPNQWPPEISRLCISVSPHRRIVRPLFLPQSSLFSSFLLPPKQMNNGTAADLRSHSLVQCKPIWDLPHQQTLVDKWAPGLSRKSPSPWQSRFPLQDGRVLAVSTHFVSDHSEEQSPCHTLFYDFALLTVGADNSSKTTKCARQRSSGWFFANARNVLAFPYFWACSPSISIRKGTLINCQSLTKH
>g4 1 9212 7856 - 000085K
MNAFCSAPSEGPVTIELRQKIVDFFKPEHLEVECESRLHNVPKGAEKHFRVQIVSDKFNGMTQLHRQRMVNKLLAEELREKIHALRIEAKIPSEWHGTKQTPAPPCEGGEKKRRQSEQPKRQ
>g5 1 14129 11906 - 000085K
MLSPLASSSSARPSYSRHHKQSTYFHRPTLIKSEPEELCSSVPPVSSLSSSSSLSSSSSSSSSSLSSSSLSSSSKSLVPKQLRPPPLIDTSTPESLERDFHAWLKGELSRSERPFCARHSDCFSAQSPPPSAMFLTPLGNVKNELVVCPTDFGGDFVFGMELEPSNSDFCASTAAFAPSISMNGPFPPMQTQMIRMPMPSLSASITQEDQQMAYLLSSAGQSPSGSSQHSSLEGSPLQPNCHQTMLLSIGKYELNDRKMLTELSGEVKTDGAKKMAATDAAMTAPNSPGGRSSGTNQLGRTYSPGLPLSMVERQKIVQLFHEGWKICDISKYLCVTHSCVSKILQRFRATGSVRPKDAKEGRQESPLVAAIRDYRQRLGIMRQSEIREQLIRDGLCRRENAPSRSSINHILRTKLSGLEAPTIGKRSSNQTTENANGKRQTDGRGENDGTAANAATARVYYDE
etc.
operon.db
1
2
3
4
5
6
7
8
9
>GPLIN_000000100 1 1060 32 + 1
VGSAGLLCETGGFCPWLRAVSLRGGAWTSGESCLWSASAACMLVRAGVRQESGLSSHARKLHSSEKEARANLHAPRCLSAMPKFTATFEANTLSELLSQIKGVLAEGNSVGVGAKKPSVGEAGGVAKAGGKAAKRKRLRQQAAAAVVAAKHAKGGDEEAAETSEKQVDPSSTKPPKKDKPHRKSLEGKE
>GPLIN_000000200 1 48028 47003 + 1
MSKAAEPLLKDAIISLGPPNSTIVCAVSGHPTSFPQRLCNAIFSKIYELGITILVSKLEGRFLHVQLGSGMDALMALSMDGVVLGEDLRLEVCLGFAGCWLDLLMADLQCLGVVREIRMRKVMNCLAMGKRLFWTMFRSLISPM
>GPLIN_000000300 1 67668 48851 - 1
MADQQQQQQDDLGPPPDPVMEVPPPKEIKVLESADDIQSRRTEVLDHYAQFQGFATIKRERLEEARQFQYFKRDADELEIWILEKLQTAAEESFRDPTNLQAKIQKHEAFVAEVQAHSNAITKLDKTGNDMIQHDHYEKDTIRKRLDRLHELWDRLFAMLEGKGIKLQQTLKLLQFVRKCDEMLYWIRDKIQFVSSDDLGADLEHVEVMQRKFDEFLKELKSHESRVLDINHEANTLIDEGHPEQQQIHGKRDEVNEAWHKLGTLTSTRGEALFGAQQIQRFYRDIDETLAWMGEKELTLATDDYGRDLNNVQALQRKHEGTERDLAALESKMDKLGSETNRLCQLYPEKSGDIATKMDEAKGRWEALKLGAEQRKRGLDRSYNRHRFMADYRELGEWIKGIQALIQSSELAKDVAGAEALLEQHQEHKGEIEARADSFKQTAETGQRLLDEEDIDNAXXXXXXXXXXXXXXXXXXXXXXNLISFLATGAKLQEANQEQLFNRNIEDVELWLSELEGQLASEDFGKDLVSVQNLQKKLGLVESDYNAHQERIDAIGHQTREFHDTGHFNAPLISKKFDVLQQRFEALREPLQRRKQKLAESLLGHQLFRDIDHELAWIREKEQIAASTNRGRDLVGVQNLIKKQNALHTEIANHEQQMEQVAQSAEQMVQRGHFLAPDIRDKKSQLRDNWRALKTKADKRKQDLQDSLQAHQYLADANEAESWMRDKEPLVGSTDYGKDEDSAESLLKKHRALMSDLEAFNTTIEELRKEAGQCKYQEQLGGQLGKECVVALYEYTEKSPREVSMKKGDVLTLLNASNKDWWKVEVSDRQGFVPAAYVKRIEPGSQHHHQAMAGQPMNTINAKQSQSEDQYKRLLMLGEQRKRKLEEACKAYQLLREANDLAEWIRSRETVASQQEIGQDLEQVEVLQNKFNDFKNDLKANEVRLQEMNQIATRLSQMGQTETAVRIRQQIDDLNARWKALEQKAEERGQQLESAHEVQRFHKDVDETKDWIQEKNETLDSEDFGRDLRSVQALQRKHEGVERDLAALGDKIRALDEKANRLRQTHPEAAEQIYDLQRQLNEQWTTLTQKANRRKDKLLDRPCVSMKKGDVLTLLNASNKDWWKVEVSDRQGFVPAAYVKRIEPGSQHHHQAMAGQPMNTINAKQSQSEDQYKRLLMLGEQRKRKLEEACKAYQLLREANDLAEWIRSRETVASQQEIGQDLEQVEVLQNKFNDFKNDLKANEVRLQEMNQIATRLSQMGQTETAVRIRQQIDDLNARWKALEQKAEERGQQLESAHEVQRFHKDVDETKDWIQEKNETLDSEDFGRDLRSVQALQRKHEGVERDLAALGDKIRALDEKANRLRQTHPEAAEQIYDLQRQLNEQWTTLTQKANRRKDKLLDSYGQCFLKAALIEKRSKLGESQTLQQFSRDAERWRIGWRRSSKSRRRRPTGPTNIQQKHQKQQAFEAELSANADRIATLISAGQNLITAAKCGGGETAVSARLRALNDQWEQLVKTTTEKSHRLKEANKQKTFMAAVKDLEFWLGEVETLLASEDYGRDLASIENLLKKHQLLEADITAHADRVEDMNRQADALLESEQFDQAQIDGRRRNINERYERVKDAANVRRDKLNKAITVHQFLRDIDEEESWIKEKKLLVSSDDYGRDLAGVQNLRRKHRRFDTELATHQPQVQVVRSKGMELMASSEIGVPEIVKRIKALDQSWTHMVELTEDRHKKLQESEEFQNFIGRVEEEEAWLNEKQQILSSPNVGENMAAVQGLLKKHDTFEVDLQMHQQRIDELQRQGEELINAGNHHAKNIGTRMEQLVKHLILVKELAVRRLQKLRDNNAYMQFMWKCDVVESWIGDKEPHVRSTDFGRDLSSVQLLLNKQDTFDNGLNNFEHEGIQRVTELKDQLLEAEHEQSAAIEQRHQAVIQRWQQLLMNSLERRKKLTEAQAHFKNIEDLYLTFAKKASAFNSWFENAEEDLTDPVRCNSLEEIREHSTVGLAQAWDQLDQLAMRMQHNLEQQIQARNQSGVSEEALREFSMMFRHFDREKLGRLDHQQFKSCLRALGYDLPMVDEGQPEPEFQRILDLVDPNRDGYVTLQEFMAFMINKETENVRSSEEIEMAFRALSKEFRPYVIAEELFANLTPEQADYCINRMKPYVDAASGRTIAGALDFEQFVQSVFMN
>GPLIN_000000400 1 74810 70588 - 1
MITTRLSTCHCTFVGGFFRSHRSENFKKINKAVNIKCQPRMPKKLLPVTSELKTHFEILLERFLDQKYSQAYLNLFTLYLKTFKQFTAFSEEQAIEALAKLEDFFTKMSLDSEIQLSDLNAQLLTLLKNYDSDERHSWEVSELEQLALYTKHITGNVSDWIQNELEDAWPDELSRAAKNALGIEQISDPYAYNKTAHIIKDAVSSRGVPGFIYTILVSLDWDPIANFWYKCNPDYCFPVKGFHGINFVVLRHEIGSQQRAEQAEKWFEAQKSAMIKTIRDNDKISDLGSLIRKGAKVRRKLEEFAH
etc.
OrthologousPairs.list (tab delimited)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
GPLIN_000000100 g12170
GPLIN_000000200 g11691
GPLIN_000000300 g16041
GPLIN_000000400 g29333
GPLIN_000000500 g21617
GPLIN_000000600 g1861
GPLIN_000000700 g3508
GPLIN_000000800 g17807
GPLIN_000000900 g25809
GPLIN_000001000 g2058
GPLIN_000001100 g17530
GPLIN_000001200 g23779
GPLIN_000001300 g18358
GPLIN_000001400 g5028
GPLIN_000001500 g18095
GPLIN_000001600 g23645
GPLIN_000001700 g25395
etc
We now need to create the files necessary for running iadhore. The easiest way to do this is to create a new folder with the above three files, and run the commands below.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
mkdir subject
mkdir query
cd query/
#this creates a file for each scaffold containing gene and strand information
tr "\n" "\t" <../operon.db |sed 's/>/\n>/g'|sed 's/@/\t/g' |awk '{if ($7=="C") { print $1"+",$8}else {print $1"-",$8}}'|sed 's/>//g' |awk '{print >> $2 ".lst"; close($2)}'
#gets rid of the extra column
sed -i 's/ .*//g' *.lst
#puts all files in this directory listed in input.txt
ls *lst >input.txt
#Creates the query file for iadhore, scaffold \t folder/file.lst
paste <(cut -f 1 -d "." input.txt) <(awk '{print "query/"$1}' input.txt)>query.ini
#repeat the same procedure for the subject
cd ../subject/
tr "\n" "\t" <../genome.db |sed 's/>/\n>/g'|sed 's/@/\t/g' |awk '{if ($7=="C") { print $1"+",$8}else {print $1"-",$8}}'|sed 's/>//g' |awk '{print >> $2 ".lst"; close($2)}'
sed -i 's/ .*//g' *.lst
ls *lst >input.txt
paste <(cut -f 1 -d "." input.txt) <(awk '{print "subject/"$1}' input.txt)>subject.ini
#now we make the actual input file for iadhore which points to all of these files.
cd ../
cat query/query.ini subject/subject.ini |tr "\t" " " >iadhore.ini
Now you have the input file for iadhore that needs some manual modifications to work. Currently it looks as such.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
1 query/1.lst
2 query/2.lst
3 query/3.lst
4 query/4.lst
5 query/5.lst
6 query/6.lst
7 query/7.lst
8 query/8.lst
9 query/9.lst
10 query/10.lst
etc.
9183 query/9183.lst
9185 query/9185.lst
9186 query/9186.lst
9192 query/9192.lst
000001 subject/000001.lst
1syntmer subject/1syntmer.lst
000002 subject/000002.lst
2syntmer subject/2syntmer.lst
000003 subject/000003.lst
3syntmer subject/3syntmer.lst
4syntmer subject/4syntmer.lst
5syntmer subject/5syntmer.lst
6syntmer subject/6syntmer.lst
7syntmer subject/7syntmer.lst
000008 subject/000008.lst
8syntmer subject/8syntmer.lst
etc.
We need to add some relevant information to change this file for iadhore
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
#this is what is put in the iadhore.ini file
genome=G.pallida
1 query/1.lst
2 query/2.lst
3 query/3.lst
4 query/4.lst
5 query/5.lst
6 query/6.lst
7 query/7.lst
8 query/8.lst
9 query/9.lst
10 query/10.lst
etc...
#empty newline
genome=H.glycines
000001 subject/000001.lst
1syntmer subject/1syntmer.lst
000002 subject/000002.lst
2syntmer subject/2syntmer.lst
000003 subject/000003.lst
3syntmer subject/3syntmer.lst
4syntmer subject/4syntmer.lst
#empty newline
prob_cutoff=0.001
anchor_points=3
number_of_threads=16
visualizeAlignment=true
blast_table= OrthologousGenes.list
output_path= output
alignment_method=gg2
gap_size=15
cluster_gap=20
level_2_only=true
q_value=.05
The last step is to run iAdhore, which takes minutes
1
i-adhore iadhore.ini
Filtering of orthologs for improved synteny and ortholog calling.
If synteny exists between the species you’ve analyzed and the formatting and filtering is correct for iadhore, you should have multiplicons (syntenic regions) in your output/multiplicons.txt file. However, the accuracy of the orthologs and synteny will improve when if various filtering parameters are tested on the NWS score and the relative protein length.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
#remember this command form earlier? You can modify the parameters for which the orthologs were selected to improve synteny and ortholog calls. Be sure to check the length of your mulitiplicons also. More is not always
#this gave me 96 multiplicons with a mean size of 9 (min 3, max 29)
less iadhorePlazaTable |cut -f 2- |sed 's/CO List of similar genes with operon //g' |sed 's/://g' |grep -A 1 "GPLIN" |sed 's/--//g' |sed '/^$/d' |awk '{if(NF>1 && $4>40 && $7>.8 && $7<1.2) {print $2} else if (NF==1){print "#"$0}}' |grep "g" -B 1 |sed 's/--//g' |sed '/^$/d' |tr "\n" " " |tr "#" "\n" |sed '/^$/d' |sed 's/@/\t/g' |awk '{print $1"\t"$4}' >OrthologousGenes.list
#this gave me 80 multiplicons with a mean size of 10 (min 3, max 29)
#this is the one that I went forward with.
less iadhorePlazaTable |cut -f 2- |sed 's/CO List of similar genes with operon //g' |sed 's/://g' |grep -A 1 "GPLIN" |sed 's/--//g' |sed '/^$/d' |awk '{if(NF>1 && $4>50 && $7>.8 && $7<1.2) {print $2} else if (NF==1){print "#"$0}}' |grep "g" -B 1 |sed 's/--//g' |sed '/^$/d' |tr "\n" " " |tr "#" "\n" |sed '/^$/d' |sed 's/@/\t/g' |awk '{print $1"\t"$4}' >OrthologousGenes.list
#this gave me 61 multiplicons with a mean size of 9 (min 3, max 27)
less iadhorePlazaTable |cut -f 2- |sed 's/CO List of similar genes with operon //g' |sed 's/://g' |grep -A 1 "GPLIN" |sed 's/--//g' |sed '/^$/d' |awk '{if(NF>1 && $4>60 && $7>.8 && $7<1.2) {print $2} else if (NF==1){print "#"$0}}' |grep "g" -B 1 |sed 's/--//g' |sed '/^$/d' |tr "\n" " " |tr "#" "\n" |sed '/^$/d' |sed 's/@/\t/g' |awk '{print $1"\t"$4}' >OrthologousGenes.list
less iadhorePlazaTable |cut -f 2- |sed 's/CO List of similar genes with operon //g' |sed 's/://g' |grep -A 1 "GPLIN" |sed 's/--//g' |sed '/^$/d' |awk '{if(NF>1 && $4>60 && $7>.8 && $7<1.2) {print $2} else if (NF==1){print "#"$0}}' |grep "g" -B 1 |sed 's/--//g' |sed '/^$/d' |tr "\n" " " |tr "#" "\n" |sed '/^$/d' |sed 's/@/\t/g' |awk '{print $1"\t"$4}' >OrthologousGenes.list
#this gave me 39 multiplicons
less iadhorePlazaTable |cut -f 2- |sed 's/CO List of similar genes with operon //g' |sed 's/://g' |grep -A 1 "GPLIN" |sed 's/--//g' |sed '/^$/d' |awk '{if(NF>1 && $4>70 && $7>.7 && $7<1.3) {print $2} else if (NF==1){print "#"$0}}' |grep "g" -B 1 |sed 's/--//g' |sed '/^$/d' |tr "\n" " " |tr "#" "\n" |sed '/^$/d' |sed 's/@/\t/g' |awk '{print $1"\t"$4}' >OrthologousGenes.listwith a mean size of 8 (min 4, max 49)
less iadhorePlazaTable |cut -f 2- |sed 's/CO List of similar genes with operon //g' |sed 's/://g' |grep -A 1 "GPLIN" |sed 's/--//g' |sed '/^$/d' |awk '{if(NF>1 && $4>70 && $7>.7 && $7<1.3) {print $2} else if (NF==1){print "#"$0}}' |grep "g" -B 1 |sed 's/--//g' |sed '/^$/d' |tr "\n" " " |tr "#" "\n" |sed '/^$/d' |sed 's/@/\t/g' |awk '{print $1"\t"$4}' >OrthologousGenes.list
While a visualization of the synteny is available from the output of iadhore, it is in a scaffold by scaffold context. Here is the longest multiplicon from the above work.
With each box representing a gene, colored boxes indicate orthologues, and black boxes are genes without homology
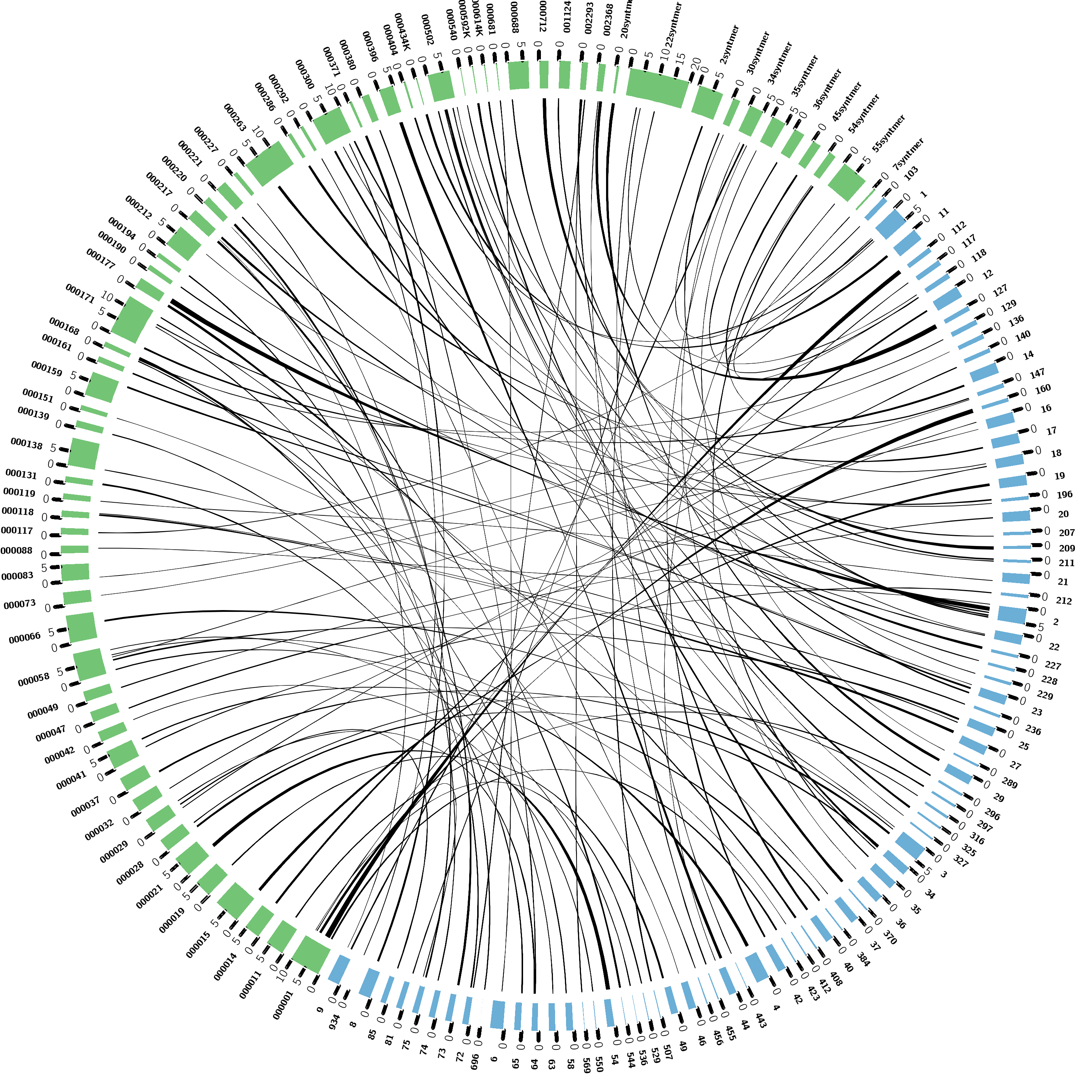
Visualization of synteny with circos
Circos is a genomic visualization tool that can display many types of data, but it is one of the most appealing and useful ways to visualize synteny. A plethora of tutorials, examples, and documentation can be found on their website, http://circos.ca/.
There are a number of files that are needed to make a circos figure in addition to the multiplicons.txt file generated above. Because circos is essentially its own code, it is easier to use templates of existing files to create them from the beginning.
There are 5 essential files.
ticks.conf
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
show_ticks = yes
show_tick_labels = yes
<ticks>
radius = 1r
color = black
thickness = 10p
multiplier = 1e-5
format = %d
<tick>
spacing = 5u
size = 25p
show_label = yes
label_size = 25p
label_offset = 10p
format = %d
</tick>
</ticks>
bands.conf
1
2
3
4
5
<bands>
show_bands = yes
fill_bands = yes
band_transparency = 4
</bands>
ideogram.conf
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
<ideogram>
<spacing>
default = 0.006r
break = 30u
axis_break_at_edge = yes
axis_break = yes
axis_break_style = 2
<break_style 1>
stroke_color = black
thickness = 0.45r
stroke_thickness = 2p
</break>
<break_style 2>
stroke_color = black
stroke_thickness = 5p
thickness = 4r
</break>
</spacing>
radius = 0.89r
thickness = 80p
fill = yes
stroke_color = white
stroke_thickness = 4p
fill_color = black
show_label = yes
label_font = bold
label_size = 20
label_parallel = no
label_radius = dims(ideogram,radius_outer) + 0.06r
</ideogram>
circos.conf
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
karyotype = ./karyotype.conf
chromosomes_units = 100000
<<include ideogram.conf>>
<<include ticks.conf>>
<<include bands.conf>>
<links>
<link>
file=SyntenicRibbons.conf
radius = 0.98r
bezier_radius = 0.1r
thickness = 1
ribbon = yes
</link>
</links>
<image>
<<include /shared/software/GIF/programs/circos/0.69.2/etc/image.conf>>
angle_offset* = -46
</image>
<<include /shared/software/GIF/programs/circos/0.69.2/etc/colors_fonts_patterns.conf>>
<<include etc/housekeeping.conf>>
Two files to make, karyotype.conf and syntenicribbons.conf
1
2
3
4
We are going to extract the genomic positions of the first and last gene in each syntenic fragment.
To do this we need to modify the gff files to allow for perfect matches full word matches with grep -w
awk '$3=="gene"' 738.polished.mitofixed.repmod.gff3 |sed 's/;//g' |sed 's/ID=//g' >SCNMod.gff
awk '$3=="gene"' globodera_pallida.PRJEB123.WBPS7.annotations.gff3 |sed 's/;/\t/g' |sed 's/ID=gene://g' >GpalMod.gff
Now we are going to use our segments.txt file that iadhore created to create the syntenic ribbons for circos. It should look something like this.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
id multiplicon genome list first last order
1 1 H.glycines 6syntmer g10101 g10117 0
2 1 G.pallida 11 GPLIN_000069900 GPLIN_000071600 1
3 2 H.glycines 000292 g19603 g19628 0
4 2 G.pallida 54 GPLIN_000273400 GPLIN_000275600 1
5 3 G.pallida 127 GPLIN_000497100 GPLIN_000499800 0
6 3 H.glycines 53syntmer g14365 g14385 1
7 4 H.glycines 000227 g7191 g7214 0
8 4 G.pallida 29 GPLIN_000162900 GPLIN_000164500 1
9 5 G.pallida 507 GPLIN_001087900 GPLIN_001088600 0
10 5 H.glycines 000265 g7523 g7530 1
11 6 G.pallida 160 GPLIN_000577800 GPLIN_000579000 0
12 6 H.glycines 000181 g6403 g6415 1
13 7 G.pallida 49 GPLIN_000254000 GPLIN_000254600 0
14 7 H.glycines 000414K g14871 g14877 1
15 8 H.glycines 000670K g16610 g16619 0
16 8 G.pallida 40 GPLIN_000213800 GPLIN_000214600 1
17 9 G.pallida 147 GPLIN_000547400 GPLIN_000548300 0
18 9 H.glycines 000101 g9167 g9179 1
Essentially the first part of these four scripts are swapping the above columns so that G. pallida genes are always in columns 1, 2, 3, and 4 and H. glycines are in 5, 6, 7, 8, while ditching some irrelevant information.
1
less segments.txt |awk 'NR>1' |awk '{if(NR%2) {print "#"$3,$4,$5,$6}else {print $3,$4,$5,$6}}' |tr "\n" "\t" |tr "#" "\n" |awk '{if($5=="G.pallida") {print $5,$6,$7,$8,$1,$2,$3,$4} else {print $1,$2,$3,$4,$5,$6,$7,$8}}' |less
The above script is part of the four scripts below that extract the the four gene positions.
1
2
3
4
5
6
7
less segments.txt |awk 'NR>1' |awk '{if(NR%2) {print "#"$3,$4,$5,$6}else {print $3,$4,$5,$6}}' |tr "\n" " " |tr "#" "\n" |awk '{if($5=="G.pallida") {print $5,$6,$7,$8,$1,$2,$3,$4} else {print $1,$2,$3,$4,$5,$6,$7,$8}}' |awk '{print $3}' |sed '/^$/d' |while read line; do grep -w $line GpalMod.gff; done |awk '{if($7=="+") {print $5} else {print $4}}' >Col3
less segments.txt |awk 'NR>1' |awk '{if(NR%2) {print "#"$3,$4,$5,$6}else {print $3,$4,$5,$6}}' |tr "\n" " " |tr "#" "\n" |awk '{if($5=="G.pallida") {print $5,$6,$7,$8,$1,$2,$3,$4} else {print $1,$2,$3,$4,$5,$6,$7,$8}}' |awk '{print $4}' |sed '/^$/d' |while read line; do grep -w $line GpalMod.gff; done |awk '{if($7=="+") {print $4} else {print $5}}' >Col4
less segments.txt |awk 'NR>1' |awk '{if(NR%2) {print "#"$3,$4,$5,$6}else {print $3,$4,$5,$6}}' |tr "\n" " " |tr "#" "\n" |awk '{if($5=="G.pallida") {print $5,$6,$7,$8,$1,$2,$3,$4} else {print $1,$2,$3,$4,$5,$6,$7,$8}}' |awk '{print $7}' |sed '/^$/d' |while read line; do grep -w $line SCNMod.gff; done |awk '{if($7=="+") {print $5} else {print $4}}' >Col7
less segments.txt |awk 'NR>1' |awk '{if(NR%2) {print "#"$3,$4,$5,$6}else {print $3,$4,$5,$6}}' |tr "\n" " " |tr "#" "\n" |awk '{if($5=="G.pallida") {print $5,$6,$7,$8,$1,$2,$3,$4} else {print $1,$2,$3,$4,$5,$6,$7,$8}}' |awk '{print $8}' |sed '/^$/d' |while read line; do grep -w $line SCNMod.gff; done |awk '{if($7=="+") {print $4} else {print $5}}' >Col8
#This code uses the list from the first step to paste the scaffold names and scaffold positions in the proper orientation as in the example output below (space separated)
less segments.txt |awk 'NR>1' |awk '{if(NR%2) {print "#"$3,$4,$5,$6}else {print $3,$4,$5,$6}}' |tr "\n" "\t" |tr "#" "\n" |awk '{if($5=="G.pallida") {print $5,$6,$7,$8,$1,$2,$3,$4} else {print $1,$2,$3,$4,$5,$6,$7,$8}}' |awk '{print $2,$6}' |awk 'NR>1' |paste - Col3 Col4 Col7 Col8 |awk '{print $1,$3,$4,$2,$5,$6}' >SyntenicRibbons.conf
Your end file should be space separated and look like this.
<scaffold#>
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
11 75168 141884 6syntmer 8293 67069
54 20891 142511 000292 141506 243297
127 6950 165341 53syntmer 166571 252128
29 18344 100822 000227 40581 121935
507 27596 67643 000265 1098749 1117756
160 49531 159235 000181 636286 673554
49 183367 228811 000414K 142821 163767
40 154422 211237 000670K 54347 90354
147 87553 130708 000101 171712 207932
2 181497 230277 000534K 311054 365249
8 196946 262443 000431K 40428 110016
29 146506 295697 000421K 274687 337420
112 29325 102680 000167 308612 391812
63 82492 98132 000334 80396 115724
3 360072 381682 000673K 32550 61735
64 149917 186398 000327K 375677 403197
2 108037 151858 000538K 212320 276634
73 25222 55157 52syntmer 215368 264136
1 248579 304406 000324 36127 102477
21 307238 353867 35syntmer 26751 95044
412 62951 71632 001622K 607692 619348
438 40971 85597 000244 427997 443265
25 324014 336805 000380 217592 231019
423 15708 62803 000167 175861 202193
etc.
The last file that we need to make is that which describes the scaffolds we want to display. This subset of scaffolds can be taken from your SyntenicRibbons.conf file. To get scaffold lengths we can use bioawk, grep and the genome fastas. Make sure that the scaffold names in this file and the SyntenicRibbons.conf file match.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
module load bioawk
#G.pallidaKaryotype
bioawk -c fastx '{print $name,length($seq)}' globodera_pallida.PRJEB123.WBPS7.genomic_softmasked.fa |awk '{print "chr","-",$1,$1,"0",$2,"blue"}' |sed 's/pathogens_Gpal_scaffold_//g' >G.pallidaKaryotype.conf
#H.glycinesKaryotype
bioawk -c fastx '{print $name,length($seq)}' genome738sl.polished.mitoFixed.fa |awk '{print "chr","-",$1,$1,"0",$2,"green"}' >H.glycinesKaryotype.conf
#this just grabs the correct scaffold from the karyotype.conf files above that have a syntenic ribbon in SyntenicRibbons.conf
##Because this prints to a temp file, if you make a mistake, you need to delete the tempKaryotype.conf1 and tempKaryotype.conf2 files before rerunning the sh scripts again.
awk '{print $1}' SyntenicRibbons.conf|while read line; do echo "awk '\$3==\""$line"\"' G.pallidaKaryotype.conf >>tmpKaryotype.conf1";done >G.pallidaKaryotype.sh
sh G.pallidaKaryotype.sh
awk '{print $4}' SyntenicRibbons.conf|while read line; do echo "awk '\$3==\""$line"\"' H.glycinesKaryotype.conf >>tmpKaryotype.conf2";done >H.glycinesKaryotype.sh
sh H.glycinesKaryotype.sh
cat <(sort tmpKaryotype.conf1 |uniq) <(sort tmpKaryotype.conf2 |uniq) >karyotype.conf
karyotype.conf
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
chr - scaffold_5 scaffold_5 0 1042978 green
chr - scaffold_16 scaffold_16 0 874394 green
chr - scaffold_14 scaffold_14 0 891198 green
chr - scaffold_33 scaffold_33 0 628838 green
chr - scaffold_41 scaffold_41 0 566278 green
chr - scaffold_12 scaffold_12 0 911565 green
chr - scaffold_36 scaffold_36 0 602358 green
chr - scaffold_25 scaffold_25 0 714661 green
chr - scaffold_60 scaffold_60 0 465007 green
chr - scaffold_53 scaffold_53 0 491105 green
chr - scaffold_59 scaffold_59 0 466286 green
chr - scaffold_58 scaffold_58 0 469186 green
chr - scaffold_52 scaffold_52 0 499914 green
chr - scaffold_66 scaffold_66 0 430898 green
chr - scaffold_29 scaffold_29 0 657481 green
chr - scaffold_82 scaffold_82 0 381180 green
chr - scaffold_84 scaffold_84 0 375857 green
chr - scaffold_79 scaffold_79 0 384640 green
chr - scaffold_81 scaffold_81 0 383483 green
chr - scaffold_88 scaffold_88 0 354636 green
Want to make the circos figure even more pretty? How about reducing syntenic band overlap so they can be seen more clearly. Circos comes with an optional utilities suite that makes life easier.
1
2
3
4
5
6
7
8
wget http://circos.ca/distribution/circos-tools-0.22.tgz
tar -zxvf circos-tools-0.22.tgz
#We will use the G.pallida tmpKaryotype.conf1 file to get the scaffold names that we want grouped together.
sort tmpKaryotype.conf1 |uniq|awk '{print $3}' |tr "\n" "," |sed 's/.$//' |awk '{print "circos-tools-0.22/tools/orderchr/bin/orderchr -links SyntenicRibbons.conf -karyotype karyotype.conf - "$0" -static_rx "$0 }' |less
#Then run the command that it generates.
circos-tools-0.22/tools/orderchr/bin/orderchr -links SyntenicRibbons.conf -karyotype karyotype.conf - 103,1,11,112,117,118,12,127,129,136,140,14,147,160,16,17,18,19,196,20,207,209,211,21,212,2,22,227,228,229,23,236,25,27,289,29,296,297,316,325,327,3,34,35,36,370,37,384,40,408,412,423,42,4,443,44,455,456,46,49,507,529,536,544,54,550,569,58,63,64,65,6,696,72,73,74,75,81,85,8,934,9 -static_rx 103,1,11,112,117,118,12,127,129,136,140,14,147,160,16,17,18,19,196,20,207,209,211,21,212,2,22,227,228,229,23,236,25,27,289,29,296,297,316,325,327,3,34,35,36,370,37,384,40,408,412,423,42,4,443,44,455,456,46,49,507,529,536,544,54,550,569,58,63,64,65,6,696,72,73,74,75,81,85,8,934,9
#this program then outputs a command that will go in your circos.conf file #circos.conf now.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
karyotype = ./karyotype.conf
chromosomes_units = 100000
<<include ideogram.conf>>
<<include ticks.conf>>
<<include bands.conf>>
<links>
<link>
file=SyntenicRibbons.conf
radius = 0.98r
bezier_radius = 0.1r
thickness = 1
ribbon = yes
</link>
</links>
<image>
<<include /shared/software/GIF/programs/circos/0.69.2/etc/image.conf>>
angle_offset* = -46
chromosomes_order = 000001,160,75,117,423,112,529,2,30syntmer,001124,000286,000177,000088,000058,16,536,25,412,4,000502,325,370,22,327,296,000168,46,58,72,129,000171,37,696,14,196,000300,211,207,34,934,1,000117,000404,7syntmer,118,36syntmer,55syntmer,22syntmer,23,000614K,000161,000159,3,000037,000066,000151,000592K,000028,000029,85,64,140,000217,227,9,544,002293,40,569,443,507,34syntmer,42,36,147,000073,45syntmer,000032,18,000049,000540,000212,000712,000190,000118,44,54syntmer,000396,000380,73,000220,000227,316,000041,212,384,27,408,209,002368,455,127,103,35,11,000434K,12,000014,136,000083,17,000263,19,000015,20,000047,21,000119,228,000221,229,000688,236,000138,289,35syntmer,29,20syntmer,297,000011,456,000292,49,000019,54,000021,550,000042,63,000139,65,000131,6,000681,74,000194,81,2syntmer,8,000371
</image>
<<include /shared/software/GIF/programs/circos/0.69.2/etc/colors_fonts_patterns.conf>>
<<include /shared/software/GIF/programs/circos/0.69.2/etc/housekeeping.conf>>
This gives us a nice visualization of synteny, as well as an estimate to which the extent of synteny exists between your two species.